Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
December 23, 2022 5:15 AM
|
Developmental timing of programmed DNA elimination in Paramecium tetraurelia recapitulates germline transposon evolutionary dynamics
At the crossroads of two different time-scales: how mechanistic constraints on somatic DNA elimination during development have streamlined Paramecium transposon-related sequences in the germline during evolution. Transposable elements (TEs) have colonized the genomes of all living organisms. Because TE integration events disrupting coding sequences can severely compromise host fitness or survival, they have generally been counter-selected, especially in the germline. With its nuclear dimorphism, the ciliate Paramecium provides a powerful unicellular model to study how eukaryotic genomes cope with TE invasion. Indeed, its germline genome, hosted in the transcriptionally silent micronuclei (MIC), has been invaded by numerous TEs and TE-related sequences, including inside genes. Gene expression takes place in a polyploid somatic macronucleus (MAC) that is destroyed at each sexual cycle, while a new MAC is formed from a copy of the MIC. New MAC development involves extensive DNA amplification, massive TE elimination and the precise excision of thousands of short TE-derived sequences called Internal Eliminated Sequences (IESs). Programmed DNA elimination is mainly guided by non-coding RNAs and repressive chromatin marks.
To gain insight into how Paramecium IESs are targeted for elimination, the “Programmed genome rearrangements” team embarked on a genome-wide study of the developmental timing of DNA elimination, in collaboration with the Cytometry and Sequencing platforms of I2BC. By combining fluorescence-assisted nuclear sorting with high-throughput DNA sequencing, they established the developmental time-course of DNA elimination at unprecedented resolution. They showed that IESs are excised following a sequential order that reflects their evolutionary age. The most ancient elements have evolved in optimizing their excision efficiency, acquiring strong sequence determinants and escaping epigenetic control. More information: https://genome.cshlp.org/content/early/2022/11/22/gr.277027.122.abstract Contact: Vinciane Regnier <vinciane.regnier@i2bc.paris-saclay.fr> & Mireille Bétermier <mireille.betermier@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 19, 2022 10:23 AM
|
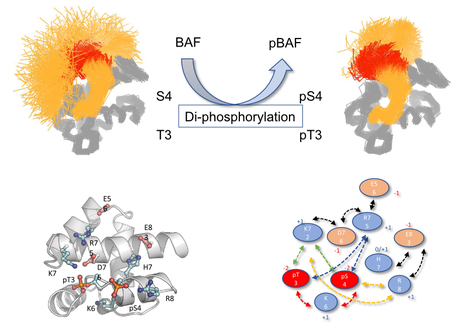
How Molecular Dynamics simulation and NMR show the effect of phosphorylation and pH on the conformational regulation of the intrinsically desordered N-terminal region of Barrier-to-Integration fact...
Mono- and, to a larger extent, di-phophorylation contribute to a gradual restriction of the conformationnal dynamics in the N-terminal region of the Protein BAF: A general mechanism for protein phosphorylation ? We studied the consequences of phosphorylation (the most frequent post-translational modification) on the conformation and dynamics of Barrier-to-Autointegration Factor (BAF) by numerical simulation and NMR. BAF is a highly conserved DNA binding protein, important for genome integrity. Its localization and function are regulated by phosphorylation. Previously published structures of BAF suggested that it is fully ordered, but our previous NMR analysis revealed that its N-terminal region is flexible in solution and that di-phosphorylation of residues S4 and T3 by the VRK1 kinase reduces this flexibility. Thanks to the computational means of TGCC via GENCI (Très Grand Centre de Calcul du CEA), we designed a molecular dynamics (MD) simulation protocol with multiple long duration (microsecond) simulations, each trajectory being characterized by the convergence of the conformational entropy of the simulated protein to ensure the completeness of the analyzed sets. This robust protocol allowed us to define the conformational ensembles accessible to the N-terminal region of BAF, whether unphosphorylated, mono-phosphorylated on S4, or di-phosphorylated on S4 and T3, and to reveal the interactions that help define these ensembles. We showed that the intrinsic flexibility of the N-terminal region of BAF is reduced by phosphorylation of the S4 residue and to a greater extent by di-phosphorylation of S4 and T3 residues. Thanks to the atomic description offered by MD simulation supported by by the NMR study of several BAF mutants, we detailed the mechanics of this restriction and described precisely the dynamic network of interactions responsible for the conformational restriction involving the phosphorylated residues pS4 and pT3. We also showed that the flexibility of the N-terminal region of BAF is influenced by ionic strength, pH and the essential role of the two negative charges carried by the phosphoryl groups for a substantial decrease in flexibility. Thus, the conformation of the intrinsically disordered N-terminal region of BAF appears to be highly adjustable, probably in connection with its various functions. This study also shows the contribution of numerical simulation in the context of structural biology for the study of the effect of post-translational modifications on protein structure and dynamics. More information: https://doi.org/10.1016/j.jmb.2022.167888 Contact: Philippe Cuniasse <philippe.cuniasse@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 2, 2022 11:00 AM
|
Ribosomal RNA operons define a central functional compartment in the Streptomyces chromosome
Ribosomal RNA operon-based evolutionary history in Streptomyces: rrn operons define a functional compartment prone to transcription and are close to pericentric inversions. Towards the existence of functional compartments in bacteria? Streptomyces are prolific producers of specialized metabolites with applications in medicine and agriculture. These bacteria possess a large linear chromosome genetically compartmentalized: core genes are grouped in the central part, while terminal regions are populated by poorly conserved genes. In exponentially growing cells, chromosome conformation capture unveiled sharp boundaries formed by ribosomal RNA (rrn) operons that segment the chromosome into multiple domains. Here we further explore the link between the genetic distribution of rrn operons and Streptomyces genetic compartmentalization. A large panel of genomes of species representative of the genus diversity revealed that rrn operons and core genes form a central skeleton, the former being identifiable from their core gene environment. We implemented a new nomenclature for Streptomyces genomes and trace their rrn-based evolutionary history. Remarkably, rrn operons are close to pericentric inversions. Moreover, the central compartment delimited by rrn operons has a very dense, nearly invariant core gene content. Finally, this compartment harbors genes with the highest expression levels, regardless of gene persistence and distance to the origin of replication. Our results highlight that rrn operons are structural boundaries of a central functional compartment prone to transcription in Streptomyces. More information: here Contact: Stephanie Bury-Mone <stephanie.bury-mone@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
November 24, 2022 9:24 AM
|
Holy coli: a mouse in the odour of sanctity
An art project to pay tribute to the laboratory mouse, often sacrificed on the altar of science. More information: here Contact: Sylvie Lautru <sylvie.lautru@i2bc.paris-saclay.fr>
|

|
Scooped by
I2BC Paris-Saclay
December 19, 2022 10:25 AM
|
The 2.7 Å resolution cryo-EM structure of a bacterial virus genome gatekeeper in the post-DNA packaging state
Structure and assembly mechanism of bacteriophage SPP1 genome gatekeeper reveal the evolutionary divergence path between different tailed phage morphotypes and herpesviruses. Numerous viruses package their double-stranded DNA genome into preformed procapsids through a portal gatekeeper that is subsequently closed. In tailed prokaryotic viruses, the gatekeeper is present at the unique vertex of the icosahedral capsid that connects with the tail.
This collaborative study reports the cryo-electron microscopy (cryoEM) structure of the complete 900 kDa connector complex that controls viral genome traffic in the capsid of bacteriophage SPP1. The high resolution of the cryo-EM electron density maps allowed tracing de novo the 30 polypeptide chains that compose the connector complex. Then we compared the connector architecture with structures, previously determined, of all its individual components in an assembly-naïve state. This allowed to unravel in molecular detail the sequential program of conformational changes and protein folding events leading to assembly of the connector until its closure to retain the viral DNA inside capsids (https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-022-34999-8/MediaObjects/41467_2022_34999_MOESM4_ESM.mp4).
The SPP1 genome gatekeeper architecture and its assembly mechanism provided a framework to amalgamate structural information from different viral systems that reveals the evolutionary process of the tailed prokaryotic viruses-herpesviruses lineage. The DNA gatekeeper of the lineage has a common module, the portal protein, while its other components diverged. Their structures identify successive evolutionary breakpoints of prokaryotic viruses diversification according to their tail morphotypes and of herpes viruses. More information: https://www.nature.com/articles/s41467-022-34999-8 Contact: Paulo Tavarès <paulo.tavares@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 19, 2022 10:21 AM
|
Exploring the potential of the model cyanobacterium Synechocystis PCC 6803 for the photosynthetic production of various high-value terpenes
We performed the first-time engineering and comparative analysis of the best studied cyanobacterium Synechocystis PCC6803 for the photosynthetic production of five chemically high-value terpenes: from CO2 (biofuels, flavors) and water (even polluted). Cyanobacteria, “microalgae” that colonise most of our planet’s ecosystems, have the potential for the sustainable production of carbon compounds from solar energy, water (even polluted) and CO2. We focused on the production of terpenes, high-value, volatile compounds (no energy required to harvest and break the biomass that produces them) that have a pleasant odour and a high energetic density. Hence terpenes can be used for the production of drugs, flavors, fragrances, solvents and/or biofuels. We have performed the first-time engineering and comparative analysis of the best-studied cyanobacterium Synechocystis PCC 6803 for the photosynthetic production of five chemically diverse high-value terpenes: two monoterpenes (C10H16) limonene (cyclic molecule) and pinene (bicyclic), and three sesquiterpenes (C15H24) bisabolene (cyclic), farnesene (linear) and santalene (cyclic). All terpene producers appeared to grow well and to be genetically stable. We showed that Synechocystis PCC 6803 can efficiently and stably produce farnesene and santalene, which had never been produced before by this model organism or any other cyanobacteria, respectively. We have also shown for the first time that Synechocystis is able to produce terpenes efficiently, when nitrate, the classical nitrogen source of cyanobacteria, is replaced by urea (cheaper than nitrate) to save costs of future industrial production or to combine production and depollution. Very interestingly, our results indicate, that Synechocystis is more proficient at producing sesquiterpenes (C15H24) than monoterpenes (C10H16), for reasons we currently investigate. More information: https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-022-02211-0 Contact: Franck Chauvat <franck.chauvat@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 2, 2022 10:58 AM
|
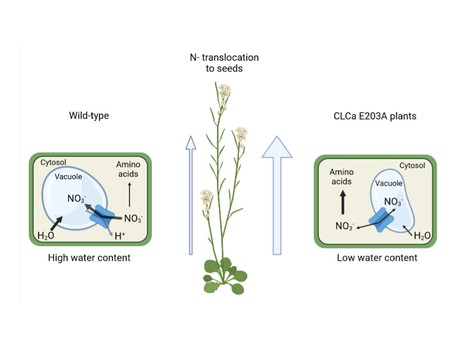
Proton exchange by the vacuolar nitrate transporter CLCa is required for plant growth and nitrogen use efficiency
CLCa exchange mechanism limits nitrogen use efficicency in plants. Nitrogen (N) is quantitatively the most important inorganic nutrient for plants. Roots mainly take it up in the form of nitrate (NO3-). Once inside the cells, nitrate can be assimilated to amino acids or stored in the vacuole, where it regulates cell water content to sustain plant growth. CLCa is the main transporter mediating the entry of NO3- into vacuoles. It works as an exchanger removing one proton from the vacuole to store two NO3- anions. CLCa belongs to a conserved family composed of both anions/protons exchangers and anions channels, with closely similar structures. Most of the exchangers share a conserved glutamate residue (E203 in CLCa). In this publication, we analyzed if the NO3-/H+ exchanger mechanism of CLCa is required for plants to stabilize water and nitrate status and what are the physiological consequences of the conversion of CLCa from an exchanger to a nitrate channel. We generated Arabidopsis lines that express a mutated form of CLCa in the amino acid E203, turning this protein into a NO3- channel. This modification decreases nitrate accumulation in the vacuole and increases amino acids and protein synthesis, thereby leading to high N content in the seeds. Although these plants present a higher nitrogen use efficiency (NUE), their growth is reduced, in association to a diminution of water content. This finding reveals the importance of the CLCa exchanger mechanism in allowing plant cells to maintain cell turgor irrespective of fluctuating nitrogen concentrations in the soil. More information: here Contact: Sophie Filleur <sophie.filleur@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
November 24, 2022 8:59 AM
|
The 6th users meeting of the The French Infrastructure for Integrated Structural Biology (FRISBI) will be held on December 9th 2022 in Gif-sur-Yvette. Preliminary program and registration: https://frisbi.eu/network/user-community/ More information: here Contact: Sophie Zinn <sophie.zinn@i2bc.paris-saclay.fr>
|




 Your new post is loading...
Your new post is loading...