 Your new post is loading...
Creating new drug candidates is a heroic endeavor, often taking over 10 years to bring a drug to market. New supercomputing-scale large language models (LLMs) that understand biology and chemistry text are helping scientists understand proteins, small molecules, DNA, and biomedical text. These state-of-the-art AI models help generate de novo proteins and molecules and predict the 3D structures of proteins. They can predict the binding structure of a small molecule to a protein and are offering scientists easier ways to engineer new candidate drugs and ultimately bring hope for patients. After Exscientia brought an AI-designed drug candidate to a clinical trial in 2021, several other companies have announced that their candidates are in trials. Within drug companies focused on AI-based discovery, there is publicly available information on about 160 discovery programs, of which 15 products are reportedly in clinical development. At the forefront of AI-based drug discovery are generative AI models for applications such as generating high-quality proteins. These large, powerful models learn from unlabeled data (such as sequencing data) on multi-GPU, multi-node, high-performance computing (HPC) infrastructures. With the novel NVIDIA BioNeMo Service, workflows for generative AI for biology are optimized and turnkey. You can focus on adapting AI models to the right drug candidates instead of dealing with configuration files and setting up supercomputing infrastructure. BioNeMo Service BioNeMo Service is a cloud service for generative AI in early drug discovery, featuring nine state-of-the-art large language and diffusion models in one place. The models in BioNeMo are accessible through a web interface or fully managed APIs and can be further trained and optimized on NVIDIA DGX Cloud. With BioNeMo Service, you can perform any of the following tasks: - Generate large libraries of proteins.
- Build property predictors using embeddings to refine protein libraries.
- Generate small molecules with specific properties.
- Rapidly and accurately predict and visualize the 3D structure for billions of proteins.
- Run large campaigns of ligand-to-small-molecule pose estimations.
- Download proteins, molecules, and predicted 3D structures.
Generative AI models in BioNeMo Service BioNeMo Service features nine AI generative models covering a wide spectrum of applications for developing AI drug discovery pipelines: - AlphaFold 2, ESMFold, and OpenFold for 3D protein structure prediction from a primary amino acid sequence
- ESM-1nv and ESM-2 for protein property predictions
- ProtGPT2 for protein generation
- MegaMolBART and MoFlow for small molecule generation
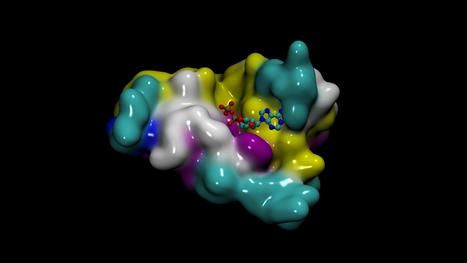
- DiffDock for predicting the binding structure of a small molecule to a protein
MIT's Kulik Lab has designed better ways of discovering and designing novel, environmentally benign, and energy-efficient materials for energy applications in a vast chemical space where molecular combinations that offer remarkable optical, conductive, magnetic, and heat transfer properties await discovery. Swift and significant gains against climate change require the creation of novel, environmentally benign, and energy-efficient materials. One of the richest veins researchers hope to tap in creating such useful compounds is a vast chemical space where molecular combinations that offer remarkable optical, conductive, magnetic, and heat transfer properties await discovery. But finding these new materials has been slow going. “While computational modeling has enabled us to discover and predict properties of new materials much faster than experimentation, these models aren’t always trustworthy,” says Heather J. Kulik PhD ’09, associate professor in the departments of Chemical Engineering and Chemistry. “In order to accelerate computational discovery of materials, we need better methods for removing uncertainty and making our predictions more accurate.” A team from Kulik’s lab set out to address these challenges with a team including Chenru Duan PhD ’22. A tool for building trust Kulik and her group focus on transition metal complexes, molecules comprised of metals found in the middle of the periodic table that are surrounded by organic ligands. These complexes can be extremely reactive, which gives them a central role in catalyzing natural and industrial processes. By altering the organic and metal components in these molecules, scientists can generate materials with properties that can improve such applications as artificial photosynthesis, solar energy absorption and storage, higher efficiency OLEDS (organic light emitting diodes), and device miniaturization. “Characterizing these complexes and discovering new materials currently happens slowly, often driven by a researcher’s intuition,” says Kulik. “And the process involves trade-offs: You might find a material that has good light-emitting properties, but the metal at the center may be something like iridium, which is exceedingly rare and toxic.” Researchers attempting to identify nontoxic, earth-abundant transition metal complexes with useful properties tend to pursue a limited set of features, with only modest assurance that they are on the right track. “People continue to iterate on a particular ligand, and get stuck in local areas of opportunity, rather than conduct large-scale discovery,” says Kulik.
AI could be just as effective in developing biochemical weapons as it is in identifying helpful new drugs, researchers warn. A group of researchers at Collaborations Pharmaceutical in North Carolina were asked by the Swiss Federal Institute for Nuclear, Biological, and Chemical Protection to present at the institute’s biannual conference on “how AI technologies for drug discovery could potentially be misused.” While not a topic they had spent much time considering, in preparation for the presentation they took their own “molecule generator” AI software and reversed the parameters: They asked it to optimize for toxicity instead of therapeutic value. In less than six hours, the software had designed 40,000 potentially toxic molecules. Among those candidates were the nerve agent VX, one of the most toxic chemical warfare agents of the 20th century, and several other known chemical warfare agents. It generated them out of thin air; none of those agents were in the dataset used to train the AI. “Without being overly alarmist, this should serve as a wake-up call for our colleagues in the ‘AI in drug discovery’ community,” the researchers wrote. Their molecule generator was based on readily available open-source tools in wide use in the pharmaceutical community; while these researchers claim they destroyed the results of their thought experiment, it would be easy for bad actors to use the same tools and public toxicity datasets to repeat their experiment and design their own novel, toxic chemicals. As the researchers point out, much of our public debate on the potential misuse of AI relates to privacy, discrimination, and safety, but not to national and international security issues such as developing new chemical and biological weapons. But the hard part here is that the tools are the same, regardless of whether they are being used to generate new life-saving drugs or new chemical warfare agents. How do you police the use of these tools so that you have one without the other? In many ways, though, this is a constantly revisited theme for technology advances. Microsoft Office and Google Docs can be used to write both hate-filled screeds and hate-crime legislation. Accounting software powers the scrappy start-up down the street as well as crime syndicates. AI-powered computer vision software can search video for wanted criminals and can surveil innocent citizens. 3D printers can make useful tools, and also unregistered, untraceable, often undetectable weapons. The most common defense we hear from the tech industry is that technology is “amoral”; the technology itself is neither good nor evil, while the uses of that technology — and the people who use them that way — are what we need to police. Easier said than done, however; in most cases where the technology is freely available, we don’t discover misuses until after the damage is done. The researchers suggest a few ideas for how to curtail some of the worst potential outcomes of the dual-nature of AI-based tools. One is to step up the development of ethical guidelines for these emerging areas of concern, followed by enhanced training for professionals on where the ethical boundaries lie. Another is to put the AI software and accompanying data models “in the cloud” where access can be regulated, rather than allowing them to be downloaded to private machines where unknown actors can use them for malevolent purposes. But in truth, as the researchers of this study put it, “the genie is out of the medicine bottle.” The tools that are already out there are enough to help someone with evil intent cause great harm. Add this to the list of challenges in a technologically advancing society: how to empower people to do good while preventing them from doing evil.
Origin-of-life researchers have proposed countless imaginative ideas about how that occurred and where the necessary raw ingredients came from. Some of the most difficult to account for are proteins, the critical backbones of cellular chemistry, because in nature today they are made exclusively by living cells. How did the first protein form without life to make it? Scientists have mostly looked for clues on Earth. Yet a new discovery suggests that the answer could be found beyond the sky, inside dark interstellar clouds. Last month in Nature Astronomy, a group of astrobiologists showed that peptides, the molecular subunits of proteins, can spontaneously form on the solid, frozen particles of cosmic dust drifting through the universe. Those peptides could in theory have traveled inside comets and meteorites to the young Earth — and to other worlds — to become some of the starting materials for life. The simplicity and favorable thermodynamics of this new space-based mechanism for forming peptides make it a more promising alternative to the known purely chemical processes that could have occurred on a lifeless Earth, according to Serge Krasnokutski, the lead author on the new paper and a researcher at the Max Planck Institute for Astronomy and the Friedrich Schiller University in Germany. And that simplicity “suggests that proteins were among the first molecules involved in the evolutionary process leading to life,” he said. Researchers say they’ve found a shortcut to proteins — a simpler chemical pathway that reenergizes the theory that proteins were present very early in the genesis of life. Whether those peptides could have survived their arduous trek from space and contributed meaningfully to the origin of life is very much an open question. Paul Falkowski, a professor at the School of Environmental and Biological Sciences at Rutgers University, said that the chemistry demonstrated in the new paper is “very cool” but “doesn’t yet bridge the phenomenal gap between proto-prebiotic chemistry and the first evidence of life.” He added, “There’s a spark that’s still missing.”
Until now, observing subatomic structures was beyond the resolution capabilities of direct imaging methods, and this seemed unlikely to change. Czech scientists, however, have presented a method with which they became the first in the world to observe an inhomogeneous electron charge distribution around a halogen atom, thus confirming the existence of a phenomenon that had been theoretically predicted but never directly observed. Comparable to the first observation of a black hole, the breakthrough will facilitate understanding of interactions between individual atoms or molecules as well as of chemical reactions, and it opens a path to refinement of the material and structural properties of various physical, biological, and chemical systems. The breakthrough discovery is being published in Science.
Functional materials are commonly employed in emerging technologies, such as green energy solutions and new electronic devices. These materials are typically blends of different organic and inorganic components and have many advantageous properties for novel applications. To achieve their full potential, we need precise knowledge on their atomic structure. State-of-the-art experimental tools, such as atomic force microscopy (AFM), can be used to investigate organic molecular adsorbates on metallic surfaces. However, interpreting the actual structure from microscopy images is often difficult. Computational simulations can help to estimate the most probable structures, but with complex materials, accurate structure search is computationally intractable with conventional methods. Recently, the CEST group has developed new tools for automated structure prediction using machine-learning algorithms from computer science. In this most recent work, researchers have demonstrated the accuracy and efficiency of the Bayesian Optimization Structure Search (BOSS) artificial intelligence method. BOSS identified the adsorbate configurations of a camphor molecule on a Cu(111) surface. This material has been previously studied with AFM, but inferring the structures from those images was inconclusive. Here, the researchers have shown that BOSS can successfully identify not only the most probable structure, but also eight stable adsorbate configurations that camphor can have on Cu(111). They used these model structures to better interpret the AFM experiments and concluded that the images likely feature camphor chemically bound to the Cu surface via an oxygen atom. Analyzing single molecular adsorbates in this way is only the first step toward studying more complex assemblies of several molecules on surfaces and subsequently the formation of monolayers. The acquired insight on interface structures could help to optimize the functional properties of these materials.
In June, the U.S. government purchased the vast majority of world’s supply of remdesivir—a FDA-approved antiviral treatment for Covid-19—for July through September. Gilead, the company that makes the compound, recently announced that it would meet international demand by the end of October. Yet all along, digital instructions for whipping up a batch of the nearly 400-atom molecule at the push of a button have been sitting on Github, an online software repository, freely available to anyone with the hardware needed to execute the chemical “program.” A dozen such chemical computers or “chemputers” sit in the University of Glasgow lab of Lee Cronin, the chemist who designed the bird’s nest of tubing, pumps, and flasks, and wrote the remdesivir code that runs on it. He’s spent years dreaming of a future where researchers can distribute and produce molecules as easily as they email and print PDFs, making not being able to order a drug as archaic as not being able to locate a modern text. Professor Lee Cronin, the lab’s principal investigator, has designed a robotic chemist called a “chemputer” that can produce chemicals from XDL programs, including the drug remdesivir, a FDA-approved antiviral treatment for the coronavirus. “If we have standard way of discovering molecules, making molecules, and then manufacturing them, suddenly nothing goes out of print,” he says. “It’s like an ebook reader for chemistry.” Cronin and his colleagues described their machine’s capability to produce multiple molecules last year, and now they’ve taken a second major step toward digitizing chemistry with an accessible way to program with the machine. Their software turns academic papers into chemputer-executable programs that researchers can edit without learning to code, they announced earlier this month in Science. And they’re not alone. The team represents one of dozens of groups spread across academia and industry all racing to bring chemistry into the digital age, a development that could lead to safer drugs, more efficient solar panels, and a disruptive new industry.
New research led by the American Museum of Natural History and funded by NASA identifies a process that might have been key in producing the first organic molecules on Earth about 4 billion years ago, before the origin of life. The process, which is similar to what might have occurred in some ancient underwater hydrothermal vents, may also have relevance to the search for life elsewhere in the universe. Details of the study are published this week in the journal Proceedings of the National Academy of Sciences. All life on Earth is built of organic molecules—compounds made of carbon atoms bound to atoms of other elements such as hydrogen, nitrogen and oxygen. In modern life, most of these organic molecules originate from the reduction of carbon dioxide (CO2) through several “carbon-fixation” pathways (such as photosynthesis in plants). But most of these pathways either require energy from the cell in order to work, or were thought to have evolved relatively late. So how did the first organic molecules arise, before the origin of life? To tackle this question, Museum Gerstner Scholar Victor Sojo and Reuben Hudson from the College of the Atlantic in Maine devised a novel setup based on microfluidic reactors, tiny self-contained laboratories that allow scientists to study the behavior of fluids—and in this case, gases as well—on the microscale. Previous versions of the reactor attempted to mix bubbles of hydrogen gas and CO2 in liquid but no reduction occurred, possibly because the highly volatile hydrogen gas escaped before it had a chance to react. The solution came in discussions between Sojo and Hudson, who shared a lab bench at the RIKEN Center for Sustainable Resource Science in Saitama, Japan. The final reactor was built in Hudson's laboratory in Maine. “Instead of bubbling the gases within the fluids before the reaction, the main innovation of the new reactor is that the fluids are driven by the gases themselves, so there is very little chance for them to escape,” Hudson said. The researchers used their design to combine hydrogen with CO2 to produce an organic molecule called formic acid (HCOOH). This synthetic process resembles the only known CO2-fixation pathway that does not require a supply of energy overall, called the Wood-Ljungdahl acetyl-CoA pathway. In turn, this process resembles reactions that might have taken place in ancient oceanic hydrothermal vents. “The consequences extend far beyond our own biosphere,” Sojo said. “Similar hydrothermal systems might exist today elsewhere in the solar system, most noticeably in Enceladus and Europa—moons of Saturn and Jupiter, respectively—and so predictably in other water-rocky worlds throughout the universe.” “Understanding how carbon dioxide can be reduced under mild geological conditions is important for evaluating the possibility of an origin of life on other worlds, which feeds into understanding how common or rare life may be in the universe,” added Laurie Barge from NASA’s Jet Propulsion Laboratory, an author on the study.
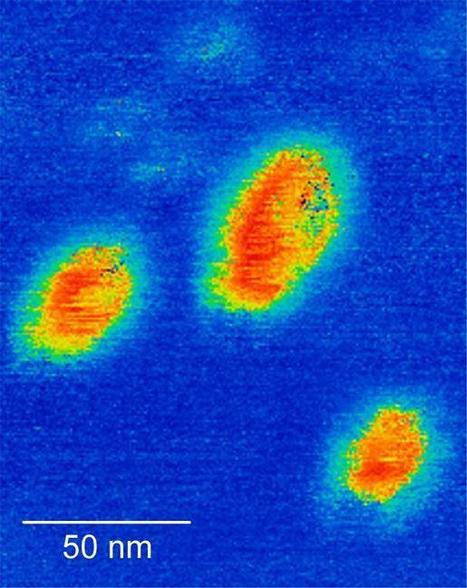
Researchers at Carnegie Mellon University have developed a method for self-assembling nanostructures with gamma-modified peptide nucleic acid (γPNA), a synthetic mimic of DNA. The process has the potential to impact nanomanufacturing, as well as future biomedical technologies like targeted diagnostics and drug delivery. Published in Nature Communications, the work introduces a science of γPNA nanotechnology that enables self-assembly in organic solvent solutions, the harsh environments used in peptide and polymer synthesis. This holds promise for nanofabrication and nanosensing. The research team, led by Rebecca Taylor, an assistant professor of mechanical engineering, reported that γPNA can form nanofibers in organic solvent solutions that can grow up to 11 microns in length (more than 1,000 times longer than their width). These represent the first complex, all-PNA nanostructures to be formed in organic solvents. Taylor, who heads the heads the Microsystems and MechanoBiology Lab at Carnegie Mellon, wants to leverage PNA’s “superpowers.” In addition to its higher thermal stability, γPNA retains the ability to bind to other nucleic acids in organic solvent mixtures that would typically destabilize structural DNA nanotechnology. This means that they can form nanostructures in solvent environments that prevent formation of DNA-based nanostructures.
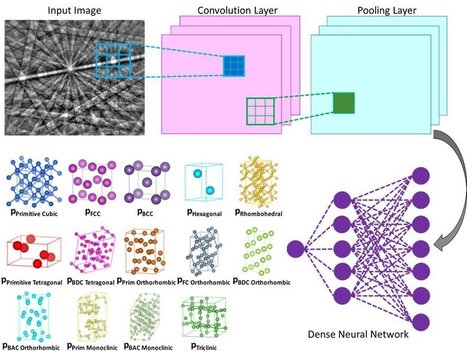
Nanoengineers at the University of California San Diego have developed a computer-based method that could make it less labor-intensive to determine the crystal structures of various materials and molecules, including alloys, proteins and pharmaceuticals. The method uses a machine learning algorithm, similar to the type used in facial recognition and self-driving cars, to independently analyze electron diffraction patterns, and do so with at least 95% accuracy. The work is published in the Jan. 31 issue of Science. A team led by UC San Diego nanoengineering professor Kenneth Vecchio and his Ph.D. student Kevin Kaufmann, who is the first author of the paper, developed the new approach. Their method involves using a scanning electron microscope (SEM) to collect electron backscatter diffraction (EBSD) patterns. Compared to other electron diffraction techniques, such as those in transmission electron microscopy (TEM), SEM-based EBSD can be performed on large samples and analyzed at multiple length scales. This provides local sub-micron information mapped to centimeter scales. For example, a modern EBSD system enables determination of fine-scale grain structures, crystal orientations, relative residual stress or strain, and other information in a single scan of the sample. However, the drawback of commercial EBSD systems is the software's inability to determine the atomic structure of the crystalline lattices present within the material being analyzed. This means a user of the commercial software must select up to five crystal structures presumed to be in the sample and then the software attempts to find probable matches to the diffraction pattern. The complex nature of the diffraction pattern often causes the software to find false structure matches in the user selected list. As a result, the accuracy of the existing software's determination of the lattice type is dependent on the operator's experience and prior knowledge of their sample. The method that Vecchio's team developed does this all autonomously, as the deep neural network independently analyzes each diffraction pattern to determine the crystal lattice, out of all possible lattice structure types, with a high degree of accuracy (greater than 95%).
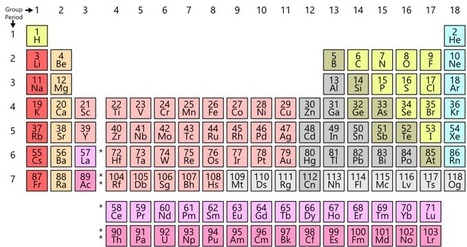
2019 is the International Year of the Periodic Table of Elements. Here, we discuss the history of the periodic table as well as the designated year. Elements 113, 115, 117 and 118 — fill out the seventh row of the periodic table of the elements. All are superheavies. That’s why they sit at the base right of the bench (see above). Naming rights classically go to those who find an element. And that’s what happen here. Element 113 was found by scientists at RIKEN in Wako, Japan. They have requested to call it Nihonium, to be shortened as Nh. This name comes from Nihon. It’s Japanese for “Land of the Rising Sun,” which is what a lot of people call Japan. Element 115 will turn out to be Moscovium, abridged as Mc. It refers to the Moscow area. And that was where the combined organization for Nuclear Research is based (Dubna). It exposed number 115 in a teamwork with researchers at Lawrence Livermore National Laboratory in California and Oak Ridge National Laboratory (ORNL) in Tennessee. That's why Tennessee also gets a periodic table entry. It’s the home location of ORNL, Vanderbilt University & the University of Tennessee. So element 117 will become Tennessine and will be represented by the symbol Ts. Russian physicist Yuri Oganessian was involved in the detection of several superheavy elements. So the group behind number 118 determined to name it after him. It becomes Oganesson — or Og. “I see it as thrilling to be familiar with that international collaborations were at the core of these discovery,” says Jan Reedijk at the Leiden organization of Chemistry in the Netherlands. He contacts the labs involved with the recently discovered elements and invite their scientists to propose names for them. Those names, Reedijk says, at the present “make the discovery somewhat tangible,” meaning seemingly more real. Element names have to follow certain rules. Allowed is: names of a scientist, a place or geographic location, a mineral, a mythological nature or concept, or some feature that is trait of the element.The recently recommended names are now open to appraisal by IUPAC and the public. Following that, the names will be official. And that’s not the finish of activities to tweak the periodic table. Physicists are already probing for still heavier elements. These would sit in a new eighth row on the table. Some scientists as well are working to confirm that copernicium is genuine. Somewhat smaller than the latest elements, it would be number 112. To evaluate all of this continuing work, chemists and physicists are collaborating to set up a latest group. They will evaluate claims of any new additions of elements.
Helium and water are known to be abundant throughout the universe, particularly in giant planets such as Uranus and Neptune. Although helium is typically nonreactive at common atmospheric conditions, past studies have found that it can sometimes react with other elements and compounds under high pressure. Researchers at Nanjing University and the University of Cambridge have recently carried out a study investigating the reaction between helium and water under high pressure conditions such as those on other planets. In their study, featured in Nature Physics, they unveiled two previously unknown types of superionic states, which they refer to as SI-I and SI-II. Superionic states are essentially phases of matter in which a compound can simultaneously exhibit both some properties of a liquid and a solid. "Helium is the most inert element in the periodic table and generally considered to be unreactive under ambient conditions," Jian Sun, one of the researchers who carried out the study, told Phys.org. "However, helium has been found react with some elements and compounds at high pressure. We wanted to understand whether helium and water can react with each other under high pressure and the nature of the states that may emerge under planetary conditions." In recent years, superionic states have become a topic of interest for many research teams worldwide. A known example of these states is superionic water (or ice), a phase of water that occurs at very high temperatures and pressures at which hydrogen atoms can move freely and oxygen atoms are fixed in their sublattice. In their study, Sun and his colleagues used calculations to show that helium (He) and water (H2O) can form several stable compounds, which exist in a wide range of pressure conditions (from 2–92 GPa). Interestingly, they found that at high pressures and temperatures, these compounds can form superionic states that have never been observed before.
In a paper released today in Science Advances, Australian researchers describe the first observation of a native ferroelectric metal: a native metal with bistable and electrically switchable spontaneous polarization states--the hallmark of ferroelectricity. The study found coexistence of native metallicity and ferroelectricity in bulk crystalline tungsten ditelluride (WTe2) at room temperature. A van-der-Waals material that is both metallic and ferroelectric in its bulk crystalline form at room temperature has potential for nano-electronics applications.
|
Catalyst materials are critical for refining petroleum products and for manufacturing pharmaceuticals, plastics, food additives, fertilizers, green fuels, industrial chemicals and much more. Catalyst materials accelerate chemical reactions without undergoing changes themselves. They are critical for refining petroleum products and for manufacturing pharmaceuticals, plastics, food additives, fertilizers, green fuels, industrial chemicals and much more. Scientists and engineers have spent decades fine-tuning catalytic reactions -- yet because it's currently impossible to directly observe those reactions at the extreme temperatures and pressures often involved in industrial-scale catalysis, they haven't known exactly what is taking place on the nano and atomic scales. This new research helps unravel that mystery with potentially major ramifications for industry. In fact, just three catalytic reactions -- steam-methane reforming to produce hydrogen, ammonia synthesis to produce fertilizer, and methanol synthesis -- use close to 10% of the world's energy. "If you decrease the temperatures at which you have to run these reactions by only a few degrees, there will be an enormous decrease in the energy demand that we face as humanity today," says Manos Mavrikakis, a professor of chemical and biological engineering at UW-Madison who led the research. "By decreasing the energy needs to run all these processes, you are also decreasing their environmental footprint." In their research, the UW-Madison engineers develop and use powerful modeling techniques to simulate catalytic reactions at the atomic scale. For this study, they looked at reactions involving transition metal catalysts in nanoparticle form, which include elements like platinum, palladium, rhodium, copper, nickel, and others important in industry and green energy. According to the current rigid-surface model of catalysis, the tightly packed atoms of transition metal catalysts provide a 2D surface that chemical reactants adhere to and participate in reactions. When enough pressure and heat or electricity is applied, the bonds between atoms in the chemical reactants break, allowing the fragments to recombine into new chemical products. "The prevailing assumption is that these metal atoms are strongly bonded to each other and simply provide 'landing spots' for reactants. What everybody has assumed is that metal-metal bonds remain intact during the reactions they catalyze," says Mavrikakis. "So here, for the first time, we asked the question, 'Could the energy to break bonds in reactants be of similar amounts to the energy needed to disrupt bonds within the catalyst?'" According to Mavrikakis's modeling, the answer is yes. The energy provided for many catalytic processes to take place is enough to break bonds and allow single metal atoms (known as adatoms) to pop loose and start traveling on the surface of the catalyst. These adatoms combine into clusters, which serve as sites on the catalyst where chemical reactions can take place much easier than the original rigid surface of the catalyst. Using a set of special calculations, the team looked at industrially important interactions of eight transition metal catalysts and 18 reactants, identifying energy levels and temperatures likely to form such small metal clusters, as well as the number of atoms in each cluster, which can also dramatically affect reaction rates. Their experimental collaborators at the University of California, Berkeley, used atomically-resolved scanning tunneling microscopy to look at carbon monoxide adsorption on nickel (111), a stable, crystalline form of nickel useful in catalysis. Their experiments confirmed models that showed various defects in the structure of the catalyst can also influence how single metal atoms pop loose, as well as how reaction sites form. Mavrikakis says the new framework is challenging the foundation of how researchers understand catalysis and how it takes place. It may apply to other non-metal catalysts as well, which he will investigate in future work. It is also relevant to understanding other important phenomena, including corrosion and tribology, or the interaction of surfaces in motion. "We're revisiting some very well-established assumptions in understanding how catalysts work and, more generally, how molecules interact with solids," Mavrikakis says.
Chemists have produced the first full quantum mechanical model of water — one of the key ingredients of life. The Journal of Physical Chemistry Letters published the breakthrough, which used machine learning to develop a model that gives a detailed, accurate description for how large groups of water molecules interact with one another. “We believe we have found the missing piece to a complete, microscopic understanding of water,” says Joel Bowman, professor of theoretical chemistry at Emory University and senior author of the study. “It appears that we now have all that we need to know to describe water molecules under any conditions, including ice, liquid or vapor over a range of temperature and pressure.” The researchers developed free, open-source software for the model, which they dubbed “q-AQUA.” The q-AQUA software provides a universal tool for studying water. “We anticipate researchers using it for everything from predicting whether an exoplanet may have water to deepening our understanding of the role of water in cellular function,” Bowman says. Bowman is one of the founders of the specialty of theoretical reaction dynamics and a leader in exploring mysteries underlying questions such as why we need water to live. First author of the study is Qi Yu, a former Emory PhD candidate in the Bowman Lab who has since graduated and is now a postdoctoral fellow at Yale. Co-authors include Emory graduate student Apurba Nandi, a PhD candidate in the Bowman Lab; Riccardo Cone, a former Emory postdoctoral fellow in the Bowman Lab, who is now at the University of Milan; and Paul Houston, former dean of science at Georgia Institute of Technology and now an emeritus professor at Cornell University. The discovery made the cover of the Journal of Physical Chemistry Letters. Water covers most of the Earth’s surface and is vital to all living organisms. It consists of simple molecules, each made up of two hydrogen atoms and one oxygen atom, bound by hydrogen. Despite water’s simplicity and ubiquity, describing the interactions of clusters of H2O molecules under any conditions presents major challenges. Newton’s law governs the behavior of heavy objects in the so-called classical world, including the motion of planets. Extremely light objects, however, at the level of atoms and electrons, are part of the quantum world which is governed by the Schrodinger equation of quantum-mechanical systems. Each water molecule consists of a single oxygen atom and two hydrogen atoms. “We’re about 70% water by weight,” Bowman says, “and yet, from a chemical standpoint, we don’t really understand how water molecules interact with biological systems." Although large, complex problems in the classical world can be divided into pieces to be solved, objects in the quantum world are too “fuzzy” to be broken down into discrete pieces. Researchers have tried to produce a quantum model of water by breaking it into the interactions of clusters of water molecules. Bowman compares it to people at a party clustered into conversational groups of two, three or four people. “Imagine you’re trying to come up with a model to describe the conversations in each of these clusters of people that can be extended to the entire party,” he says. “First you gather the data for two people talking and determine what they are saying, who is saying what and what the conversation means. It gets harder when you try to model the conversations among three people. And when you get up to four people, it gets nearly impossible because so much data is coming at you.”
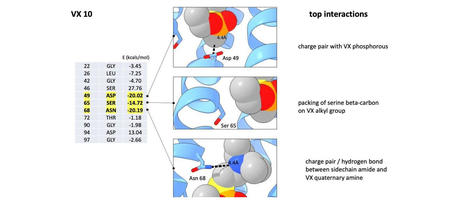
VX is classified as a neurotoxin and an incredibly deadly chemical warfare agent that has been used in assassinations by some nations. It can cause permanent brain damage in those who survive exposure. These potentially life-saving findings are published in the July 2022 edition of Science Advances, with lab member Jim McCann serving as the paper's primary author. It outlines the design of two proteins that detect the neurotoxin by changing their shape in the presence of VX. In collaboration with Douglas Pike and Vikas Nanda at Rutgers University, the CCNY team used a protein design program called ProtCAD to design 20 different proteins. According to Koder, the computer code was new and unlike anything the team had previously worked with, so it came as a bit of a surprise that two of their protein designs worked rather quickly. "Having the first thing we tried with a small molecule actually just work was pretty great," Koder said. "In that absence of VX, all of the negative charges repel each other and then the protein unfolds. And it really extends, almost like a stick. When the protein binds VX it wraps all the way around the molecule becoming much more compact." Previous detectors for this type of molecule often produced false positives from chemicals like insecticides. This new design can help prevent those misleading results, by scanning the entire molecular surface down to one hundred-millionth of a centimeter. "We get this remarkable specificity because we're making contact with the whole molecule," said Koder. This work adds to a rapidly advancing field of biosensing technology used to detect the presence of incredibly small molecules called biomarkers.
Solid-state nuclear magnetic resonance (NMR) spectroscopy—a technique that measures the frequencies emitted by the nuclei of some atoms exposed to radio waves in a strong magnetic field—can be used to determine chemical and 3D structures as well as the dynamics of molecules and materials. A necessary initial step in the analysis is the so-called chemical shift assignment. This involves assigning each peak in the NMR spectrum to a given atom in the molecule or material under investigation. This can be a particularly complicated task. Assigning chemical shifts experimentally can be challenging and generally requires time-consuming multi-dimensional correlation experiments. Assignment by comparison to statistical analysis of experimental chemical shift databases would be an alternative solution, but there is no such database for molecular solids. A team of researchers including EPFL professors Lyndon Emsley, head of the Laboratory of Magnetic Resonance, Michele Ceriotti, head of the Laboratory of Computational Science and Modeling and Ph.D. student Manuel Cordova decided to tackle this problem by developing a method of assigning NMR spectra of organic crystals probabilistically, directly from their 2D chemical structures. They started by creating their own database of chemical shifts for organic solids by combining the Cambridge Structural Database (CSD), a database of more than 200,000 three-dimensional organic structures, with ShiftML, a machine learning algorithm they had developed together previously that allows for the prediction of chemical shifts directly from the structure of molecular solids. Initially described in a Nature Communications paper in 2018, ShiftML uses DFT calculations for training, but can then perform accurate predictions on new structures without performing additional quantum calculations. Though DFT accuracy is attained, the method can calculate chemical shifts for structures with ~100 atoms in seconds, reducing the computational cost by a factor of as much as 10,000 compared to current DFT chemical shift calculations. The accuracy of the method does not depend on the size of the structure examined and the prediction time is linear in the number of atoms. This sets the stage for calculating chemical shifts in situations where it would have been unfeasible before. In the new Science Advances paper, the team used ShiftML to predict shifts on more than 200,000 compounds extracted from the CSD and then related the shifts obtained to topological representations of the molecular environments. This involved constructing a graph representing the covalent bonds between the atoms in the molecule, extending it a given number of bonds away from the central atoms. They then brought together all the identical instances of the graph in the database, allowing them to obtain statistical distributions of chemical shifts for each motif. The representation is a simplification of the covalent bonds around the atom in a molecule and doesn't contain any 3D structural features: this allowed them to obtain the probabilistic assignment of the NMR spectra of organic crystals directly from their two-dimensional chemical structures through a marginalization scheme that combined the distributions from all the atoms in the molecule.
Materials scientists can now shuffle layered compounds together, much like combining two different decks of cards. The technique, recently discovered by a team of researchers at the U.S. Department of Energy's Ames Laboratory, is leading to development of new materials with unusual electron transport properties that have potential applications in next-generation quantum technologies. The discovered technique has shown another unexpected and promising application in new materials design. The "reshuffling" approach can generate thermally stable three-dimensional (3D) heterostructures from layered transition metal dichalcogenides (TMDCs). These are van der Waals materials composed of metal nanolayers sandwiched between two other layers of chalcogens—sulfur, selenium, or tellurium. Similar to graphite, these compounds can be exfoliated into 2D layers, which display unique electron transport properties and quantum phenomena. "TMDCs are very intriguing to researchers as a possibility for applications in renewable energy, catalysis and optoelectronics, to name only a few," said project leader Viktor Balema, a Senior Scientist in the Divisions of Materials Sciences and Engineering at Ames Laboratory. "Our goal in research has been the development of such re-assembly methods for these layered materials, which are not only efficient, but also scalable and cost effective in production." Researchers at Ames Laboratory have been successful in overcoming one of the major challenges of composing these layered materials—the difficulty of sandwiching together atomically dissimilar, incommensurate, materials—through the use of mechanochemistry that is facilitated by ball milling. "Now, we have demonstrated that we can mechanochemically design novel layered heterostructures, control their composition and tune their properties," said Ihor Hlova, a scientist in the Divisions of Materials Sciences and Engineering at Ames Laboratory. "This opens a way to a variety of different combinations—the possibilities are basically unlimited."
The scientists who re-engineered the plastic-eating enzyme PETase have now created an enzyme ‘cocktail’ which can digest plastic up to six times faster. A second enzyme, found in the same rubbish dwelling bacterium that lives on a diet of plastic bottles, has been combined with PETase to speed up the breakdown of plastic. PETase breaks down polyethylene terephthalate (PET) back into its building blocks, creating an opportunity to recycle plastic infinitely and reduce plastic pollution and the greenhouse gases driving climate change. PET is the most common thermoplastic, used to make single-use drinks bottles, clothing and carpets and it takes hundreds of years to break down in the environment, but PETase can shorten this time to days. The initial discovery set up the prospect of a revolution in plastic recycling, creating a potential low-energy solution to tackle plastic waste. The team engineered the natural PETase enzyme in the laboratory to be around 20 percent faster at breaking down PET. Now, the same trans-Atlantic team have combined PETase and its ‘partner’, a second enzyme called MHETase, to generate much bigger improvements: simply mixing PETase with MHETase doubled the speed of PET breakdown, and engineering a connection between the two enzymes to create a ‘super-enzyme’, increased this activity by a further three times. The study is published in the journal Proceedings of the National Academy of Sciences of the United States of America.
The ever-increasing production and use of plastics over the last half century has created a huge environmental problem for the world. Currently, most of the 4.9 billion tonnes of plastics ever produced will end up in landfills or the natural environment, and this number is expected to increase to around 12 billion tonnes by 2050. In collaboration with colleagues at universities and institutions in the UK, China and the Kingdom of Saudi Arabia, researchers in the Edwards/ Xiao group at Oxford’s Department of Chemistry have developed a method of converting plastic waste into hydrogen gas which can be used as a clean fuel, and high-value solid carbon. This was achieved with a new type of catalysis developed by the group which uses microwaves to activate catalyst particles to effectively ‘strip’ hydrogen from polymers. The findings, published in Nature Catalysis, detail how the researchers mixed mechanically-pulverised plastic particles with a microwave-susceptor catalyst of iron oxide and aluminium oxide. The mixture was subjected to microwave treatment and yielded a large volume of hydrogen gas and a residue of carbonaceous materials, the bulk of which were identified as carbon nanotubes. This rapid one-step process for converting plastic to hydrogen and solid carbon significantly simplifies the usual processes of dealing with plastic waste and demonstrates that over 97% of hydrogen in plastic can be extracted in a very short time, in a low-cost method with no CO2 burden. The new method represents an attractive potential solution for the problem of plastic waste; instead of polluting our land and oceans, plastics could be used as a valuable feedstock for producing clean hydrogen fuel and value-added carbon products. Prof Peter Edwards said: ‘This is not good applied science, but rather good science, applied. It opens up an entirely new area of catalysis in terms of selectivity and offers a potential route to the challenge of the plastic waste Armageddon, particularly in developing countries as one route to the hydrogen economy – effectively enabling them to leap-frog the sole use of fossil fuels.
Using a new machine-learning algorithm, MIT researchers have identified a powerful antibiotic that can kill a wide range of species of pathogenic bacteria, including some that are resistant to all known antibiotics. The computer model, which can screen more than a hundred million chemical compounds in a matter of days, is designed to pick out potential antibiotics that kill bacteria using different mechanisms than those of existing drugs. "We wanted to develop a platform that would allow us to harness the power of artificial intelligence to usher in a new age of antibiotic drug discovery," says James Collins, the Termeer Professor of Medical Engineering and Science in MIT's Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering. "Our approach revealed this amazing molecule which is arguably one of the more powerful antibiotics that has been discovered." In their new study, the researchers also identified several other promising antibiotic candidates, which they plan to test further. They believe the model could also be used to design new drugs, based on what it has learned about chemical structures that enable drugs to kill bacteria. "The machine learning model can explore, in silico, large chemical spaces that can be prohibitively expensive for traditional experimental approaches," says Regina Barzilay, the Delta Electronics Professor of Electrical Engineering and Computer Science in MIT's Computer Science and Artificial Intelligence Laboratory (CSAIL). In this case, the researchers designed their model to look for chemical features that make molecules effective at killing E. coli. To do so, they trained the model on about 2,500 molecules, including about 1,700 FDA-approved drugs and a set of 800 natural products with diverse structures and a wide range of bioactivities. Once the model was trained, the researchers tested it on the Broad Institute’s Drug Repurposing Hub, a library of about 6,000 compounds. The model picked out one molecule that was predicted to have strong antibacterial activity and had a chemical structure different from any existing antibiotics. Using a different machine-learning model, the researchers also showed that this molecule would likely have low toxicity to human cells. This recently identified molecule, which the researchers decided to call halicin, after the fictional artificial intelligence system from “2001: A Space Odyssey,” has been previously investigated as possible diabetes drug. The researchers tested it against dozens of bacterial strains isolated from patients and grown in lab dishes, and found that it was able to kill many that are resistant to treatment, including Clostridium difficile, Acinetobacter baumannii, and Mycobacterium tuberculosis. The drug worked against every species that they tested, with the exception of Pseudomonas aeruginosa, a difficult-to-treat lung pathogen. To test halicin’s effectiveness in living animals, the researchers used it to treat mice infected with A. baumannii, a bacterium that has infected many U.S. soldiers stationed in Iraq and Afghanistan. The strain of A. baumannii that they used is resistant to all known antibiotics, but application of a halicin-containing ointment completely cleared the infections within 24 hours. Preliminary studies suggest that halicin kills bacteria by disrupting their ability to maintain an electrochemical gradient across their cell membranes. This gradient is necessary, among other functions, to produce ATP (molecules that cells use to store energy), so if the gradient breaks down, the cells die. This type of killing mechanism could be difficult for bacteria to develop resistance to, the researchers say. “When you’re dealing with a molecule that likely associates with membrane components, a cell can’t necessarily acquire a single mutation or a couple of mutations to change the chemistry of the outer membrane. Mutations like that tend to be far more complex to acquire evolutionarily,” Stokes says. The researchers also found that E. coli did not develop any resistance to halicin during a 30-day treatment period. In contrast, the bacteria started to develop resistance to the antibiotic ciprofloxacin within one to three days, and after 30 days, the bacteria were about 200 times more resistant to ciprofloxacin than they were at the beginning of the experiment.
Researchers at Princeton University in the US have become the first to observe a robust supercurrent at the edge of a superconductor that is very different to the supercurrent in the material’s bulk. This “topological superconductivity” could come in useful for a host of new applications, they say. Topological materials are materials that have very different properties at the surface compared to those in their bulk thanks to special topologically protected “edge states”. Topological insulators – materials that act as insulators in their interior but conduct currents on their surface – have been a hot topic in condensed-matter research for several years, but their superconducting counterparts are less well-studied. To find out what happens when the interior of a topological material is not an insulator but a superconductor, Nai Phuan Ong and colleagues turned their attention to molybdenum ditelluride (MoTe2). This material is a Weyl semimetal, a recently discovered class of topological material in which electrons (which are fermions, and thus have spin-1/2) behave as if they have no mass. These oddly-behaved particles were predicted in 1929 by theoretical physicist Herman Weyl as a solution to the Dirac equation (which describes the physics of normal fermions), and they travel faster and dissipate less energy than electrons in ordinary metals or semiconductors. They also show the “chiral magnetic effect” when placed in a magnetic field, which generates a current of positive and negative Weyl particles that move parallel and antiparallel to the field. Saw-tooth pattern in critical current oscillations The researchers began by preparing slivers of crystals of MoTe2 that were between 60 to 120 nm thick. They then cooled these crystalline samples down to below 100 millikelvin – the superconducting transition temperature of MoTe2. Next, they applied a weak magnetic field to the samples while measuring the current flow through them. The Princeton team observed a quantity known as the critical current oscillating in a saw-tooth pattern as they increase the applied magnetic field. Both the height and frequency of these oscillations fit well with predictions of how these fluctuations arise from the quantum behavior of electrons confined to the edges of the material, they say. In superconducting materials, electrons overcome their mutual electrostatic repulsion to form Cooper pairs thanks to interactions between the electrons and vibrations of the material’s crystalline lattice. Once formed, these pairs behave as bosons, which have an integer spin. This means they can condense to form a “superfluid” state that behaves as a single entity, carrying electrical current through the material with no resistance at temperatures below the material’s transition temperature.
A team of researchers at McMaster University has developed a self-cleaning surface that can repel all forms of bacteria, preventing the transfer of antibiotic-resistant superbugs and other dangerous bacteria in settings ranging from hospitals to kitchens. The new plastic surface – a treated form of conventional transparent wrap – can be shrink-wrapped onto door handles, railings, IV stands and other surfaces that can be magnets for bacteria such as MRSA and C. difficile. The treated material is also ideal for food packaging, where it could stop the accidental transfer of bacteria such as E. coli, Salmonella and listeria from raw chicken, meat and other foods, as described in a paper published today by the journal ACS Nano. The research was led by engineers Leyla Soleymani and Tohid Didar, who collaborated with colleagues from McMaster’s Institute for Infectious Disease Research and the McMaster-based Canadian Centre for Electron Microscopy. Inspired by the water-repellent lotus leaf, the new surface works through a combination of nano-scale surface engineering and chemistry. The surface is textured with microscopic wrinkles that exclude all external molecules. A drop of water or blood, for example, simply bounces away when it lands on the surface. The same is true for bacteria.
Researchers at the University of California, Irvine have developed a new scanning transmission electron microscopy method that enables visualization of the electric charge density of materials at sub-angstrom resolution. With this technique, the UCI scientists were able to observe electron distribution between atoms and molecules and uncover clues to the origins of ferroelectricity, the capacity of certain crystals to possess spontaneous electric polarization that can be switched by the application of an electric field. The research, which is highlighted in a study published today in Nature, also revealed the mechanism of charge transfer between two materials. "This method is an advancement in electron microscopy—from detecting atoms to imaging electrons—that could help us engineer new materials with desired properties and functionalities for devices used in data storage, energy conversion and quantum computing," said team leader Xiaoqing Pan, UCI's Henry Samueli Endowed Chair in Engineering and a professor of both materials science & engineering and physics & astronomy. Employing a new aberration-corrected scanning transmission electron microscope with a fine electron probe measuring half an angstrom and a fast-direct electron detection camera, his group was able to acquire a 2-D raster image of diffraction patterns from a region of interest in the sample. As obtained, the data sets are 4-D, since they consist of 2-D diffraction patterns from each probe location in a 2-D scanning area. "With our new microscope, we can routinely form an electron probe as small as 0.6 angstrom, and our high-speed camera with angular resolution can acquire 4-D STEM images with 512 x 512 pixels at greater than 300 frames per second," Pan said. "Using this technique, we can see the electron charge distribution between atoms in two different perovskite oxides, non-polar strontium titanate and ferroelectric bismuth ferrite."
When the tension rises, unexpected things can happen – not least when it comes to gold atoms. Researchers from, among others, Chalmers University of Technology, have now managed, for the first time, to make the surface of a gold object melt at room temperature. Ludvig de Knoop, from Chalmers’ Department of Physics, placed a small piece of gold in an electron microscope. Observing it at the highest level of magnification and increasing the electric field step-by-step to extremely high levels, he was interested to see how it influenced the gold atoms. It was when he studied the atoms in the recordings from the microscope, that he saw something exciting. The surface layers of gold had actually melted – at room temperature. "I was really stunned by the discovery. This is an extraordinary phenomenon, and it gives us new, foundational knowledge of gold,” says Ludvig de Knoop. What happened was that the gold atoms became excited. Under the influence of the electric field, they suddenly lost their ordered structure and released almost all their connections to each other. Upon further experimentation, the researchers discovered that it was also possible to switch between a solid and a molten structure. The discovery of how gold atoms can lose their structure in this way is not just spectacular, but also groundbreaking scientifically. Together with the theoretician Mikael Juhani Kuisma, from the University of Jyväskylä in Finland, Ludvig de Knoop and colleagues have opened up new avenues in materials science. The results are now published in the journal Physical Review Materials.
|



 Your new post is loading...
Your new post is loading...























Oxycodone without a prescription
Phentermine 37.5 mg for sale
Phentremin weight loss
purchase Adderall online
Where Fentanyl Patches online
Where to buy Acxion Fentermina