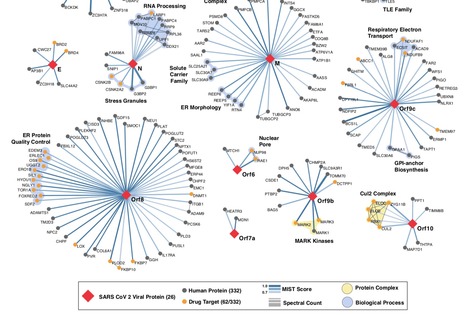
We cloned, tagged and expressed 26 of the 29 viral proteins in human cells and identified the human proteins physically associated with each using affinity-purification mass spectrometry (AP-MS), which identified 332 high confidence SARS-CoV-2-human protein-protein interactions (PPIs).
Among these, we identify 67 druggable human proteins or host factors targeted by 69 existing FDA-approved drugs, drugs in clinical trials and/or preclinical compounds, that we are currently evaluating for efficacy in live SARS-CoV-2 infection assays. The identification of host dependency factors mediating virus infection may provide key insights into effective molecular targets for developing broadly acting antiviral therapeutics against SARS-CoV-2 and other deadly coronavirus strains.
It is important to note that pharmacological intervention with the agents we identified in this study could be either detrimental or beneficial for infection. For instance, the HDAC2 inhibitors may compound the potential action of the Nsp5 protease to hydrolyze this human protein. Future work will involve generation of protein-protein interaction maps in different human cell types, as well as bat cells, and the study of related coronaviruses including SARS-CoV, MERS-CoV and the less virulent OC43..
Preprint Available at bioRXiv (March 27, 2020):
https://www.biorxiv.org/content/10.1101/2020.03.22.002386v3
Via Juan Lama


 Your new post is loading...
Your new post is loading...