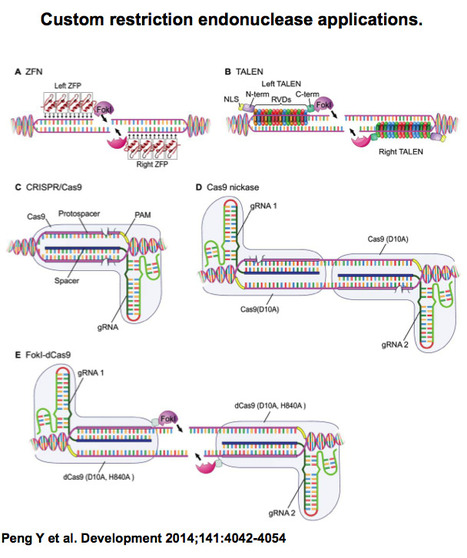
Recent advances in the targeted modification of complex eukaryotic genomes have unlocked a new era of genome engineering. From the pioneering work using zinc-finger nucleases (ZFNs), to the advent of the versatile and specific TALEN systems, and most recently the highly accessible CRISPR/Cas9 systems, we now possess an unprecedented ability to analyze developmental processes using sophisticated designer genetic tools. Excitingly, these robust and simple genomic engineering tools also promise to revolutionize developmental studies using less well established experimental organisms.
Modern developmental biology was born out of the fruitful marriage between traditional embryology and genetics. Genetic tools, together with advanced microscopy techniques, serve as the most fundamental means for developmental biologists to elucidate the logistics and the molecular control of growth, differentiation and morphogenesis. For this reason, model organisms with sophisticated and comprehensive genetic tools have been highly favored for developmental studies. Advances made in developmental biology using these genetically amenable models have been well recognized. The Nobel prize in Physiology or Medicine was awarded in 1995 to Edward B. Lewis, Christiane Nüsslein-Volhard and Eric F. Wieschaus for their discoveries on the ‘Genetic control of early structural development’ usingDrosophila melanogaster, and again in 2002 to John Sulston, Robert Horvitz and Sydney Brenner for their discoveries of ‘Genetic regulation of development and programmed cell death’ using the nematode worm Caenorhabditis elegans. These fly and worm systems remain powerful and popular models for invertebrate development studies, while zebrafish (Danio rerio), the dual frog species Xenopus laevis and Xenopus tropicalis, rat (Rattus norvegicus), and particularly mouse (Mus musculus) represent the most commonly used vertebrate model systems. To date, random or semi-random mutagenesis (‘forward genetic’) approaches have been extraordinarily successful at advancing the use of these model organisms in developmental studies. With the advent of reference genomic data, however, sequence-specific genomic engineering tools (‘reverse genetics’) enable targeted manipulation of the genome and thus allow previously untestable hypotheses of gene function to be addressed.
Via Dr. Stefan Gruenwald



 Your new post is loading...
Your new post is loading...