Skeleton/bone marrow involvement in patients with newly diagnosed Hodgkin’s lymphoma (HL) is an important predictor of adverse outcomes1. Studies show that FDG-PET/CT upstages patients with uni- or multifocal skeleton/bone marrow uptake (BMU) when iliac crest bone marrow biopsy fails to find evidence of histology-proven involvement. The general recommendation is, therefore, that bone marrow biopsy can be avoided when FDG-PET/CT is performed at staging.
Our aim was to develop an AI-based method for the detection of focal skeleton/BMU and quantification of diffuse BMU in patients with HL undergoing staging with FDG-PET/CT. The output of the AI-based method in a separate test set was compared to the image interpretation of ten physicians from different hospitals. Finally, the AI-based quantification of diffuse BMU was compared to manual quantification.
Artificial intelligence-based classification
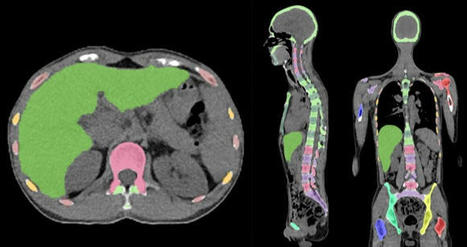
A convolutional neural network (CNN) was used to segment the skeletal anatomy11. Based on this CNN, the bone marrow was defined by excluding the edges from each individual bone; more precisely, 7 mm was excluded from the humeri and femora, 5 mm was excluded from the vertebrae and hip bones, and 3 mm was excluded from the remaining bones.
Focal skeleton/bone marrow uptake
The basic idea behind our approach is that the distribution of non-focal BMU has a light tail and most pixels will have an uptake reasonably close to the average. There will still be variations between different bones. Most importantly, we found that certain bones were much more likely to have diffuse BMU than others. Hence, we cannot use the same threshold for focal uptake in all bones. At the other end, treating each bone individually is too susceptible to noise. As a compromise, we chose to divide the bones into two groups:
-
“spine”—defined as the vertebrae, sacrum, and coccyx as well as regions in the hip bones within 50 mm from these locations, i.e., including the sacroiliac joints.
-
“other bones”—defined as the humeri, scapulae, clavicles, ribs, sternum, femora, and the remaining parts of the hip bones.
For each group, the focal standardized uptake values (SUVs) were quantified using the following steps:
- Threshold computation. A threshold (THR) was computed using the mean and standard deviation (SD) of the SUV inside the bone marrow. The threshold was set to ���=SUVmean+2SD.
- 2. Abnormal bone region. The abnormal bone region was defined in the following way:
Only the pixels segmented as bone and where SUV > THR were considered. To reduce the issues of PET/CT misalignment and spill over, a watershed transform was used to assign each of these pixels to a local maximum in the PET image. If this maximum was outside the bone mask, the uptake was assumed to be leaking into the bone from other tissues and was removed. Finally, uptake regions smaller than 0.1 mL were removed.
- 3.Abnormal bone SUV quantification. The mean squared abnormal uptake (MSAU) was first calculated as MSAU=meanof(SUV−THR)2overtheabnormalboneregion.
This calculation leads to two TSAU values; one for the “spine” and one for the “other bones”. As the TSAU value can be nonzero even for patients without focal uptake, cut-off values were tuned using the training cohort. The AI method was adjusted in the training group to have a positive predictive value of 65% and a negative predictive value of 98%. For the “spine”, a cut-off of 0.5 was used, and for the “other bones”, a cut-off of 3.0 was used. If one of the TSAU values was higher than the corresponding cut-off, the patient was considered to have focal uptake.
Results
Focal uptake
Fourteen of the 48 cases were classified as having focal skeleton/BMU by the AI-based method. The majority of physicians classified 7/48 cases as positive and 41/48 cases as negative for having focal skeleton/BMU. The majority of the physicians agreed with the AI method in 39 of the 48 cases. Six of the seven positive cases (86%) identified by the majority of physicians were identified as positive by the AI method, while the seventh was classified as negative by the AI method and by three of the ten physicians.
Thirty-three of the 41 negative cases (80%) identified by the majority of physicians were also classified as negative by the AI method. In seven of the remaining eight patients, 1–3 physicians (out of the ten total) classified the cases as having focal uptake, while in one of the eight cases none of the physicians classified it as having focal uptake. These findings indicate that the AI method has been developed towards high sensitivity, which is necessary to highlight suspicious uptake.
Conclusions
The present study demonstrates that an AI-based method can be developed to highlight suspicious focal skeleton/BMU in HL patients staged with FDG-PET/CT. This AI-based method can also objectively provide results regarding high versus low BMU by calculating the SUVmedian value in the whole spine marrow and the liver. Additionally, the study also demonstrated that inter-observer agreement regarding both focal and diffuse BMU is moderate among nuclear medicine physicians with varying levels of experience working at different hospitals. Finally, our results show that the automated method regarding diffuse BMU is comparable to the manual ROI method.
read the original paper at https://www.nature.com/articles/s41598-021-89656-9



 Your new post is loading...
Your new post is loading...