 Your new post is loading...
 Your new post is loading...
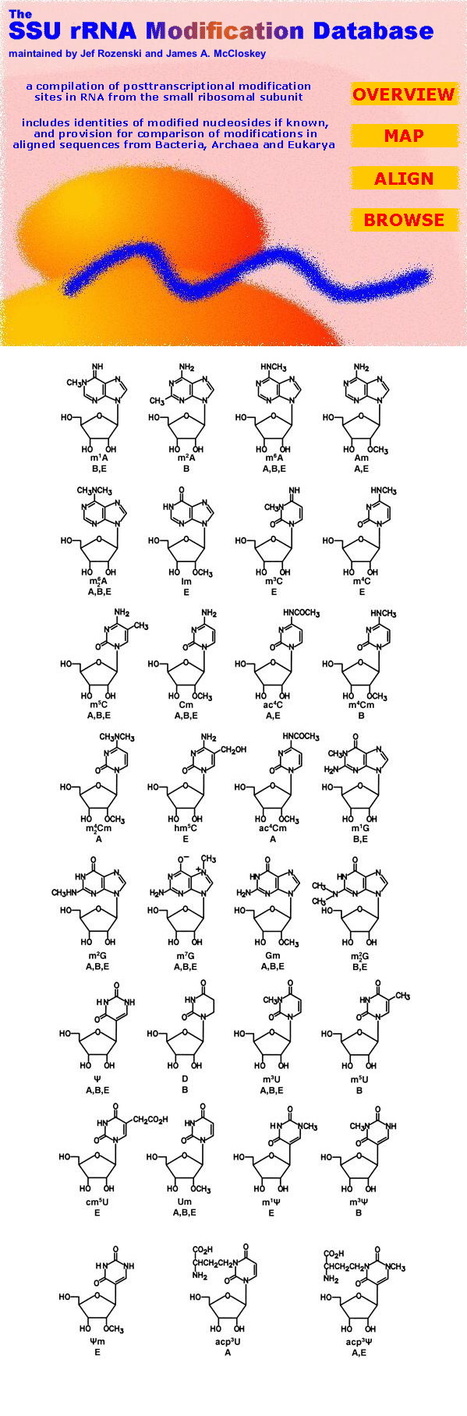
The Small Subunit rRNA Modification Database provides a listing of reported post-transcriptionally modified nucleosides and sequence sites in small subunit rRNAs from bacteria, archaea and eukarya. Data are compiled from reports of full or partial rRNA sequences, including RNase T1 oligonucleotide catalogs reported in earlier literature in studies of phylogenetic relatedness. Options for data presentation include full sequence maps, some of which have been assembled by database curators with the aid of contemporary gene sequence data, and tabular forms organized by source organism or chemical identity of the modification. A total of 32 rRNA sequence alignments are provided, annotated with sites of modification and chemical identities of modifications if known, with provision for scrolling full sequences or user-dictated subsequences for comparative viewing for organisms of interest. The database can be accessed through the World Wide Web at http://medlib.med.utah.edu/SSUmods.
In bacteria, small (~30-500 nt) non-coding RNAs (sRNAs) are the most abundant class of post-transcriptional regulators that are involved in diverse processes including quorum sensing, stress response, virulence and carbon metabolism. Based on the target molecules, sRNAs can be divided into two major groups: (i) mRNA-binding antisense sRNAs and (ii) protein-binding sRNAs. The antisense RNAs can further be categorized as cis-encoded antisense sRNAs, which are completely complementary to their targets, and trans-encoded antisense sRNAs, which are only partially complementary to their targets. In any case, the interaction between antisense RNAs and target mRNAs could direct a plethora of biological regulatory circuits. Recent developments in high-throughput techniques, such as genomic tiling arrays and RNA-Seq have provided invaluable insights into the detection and characterization of bacterial sRNAs. However, a comprehensive bacterial sRNA database is not yet available, especially for integrating and analyzing high-throughput sequencing data. Here, we have designed and constructed BSRD (Bacterial Small regulatory RNA Database) which hosts sRNAs collected from over 783 bacterial species and 957 strains. The distinctive features of BSRD are: (1) BSRD hosts sRNAs retrieved from online databases including Rfam, sRNAMap, GenBank, RegulonDB and EcoCyc, as well as manual curation.In additional, we have also integrated 20,115 regulatory elements in BSRD. (2) BSRD collects sRNAs targets predicted by computational algorithms, IntaRNA and RNAplex, as well as experimentally validated sRNAs targets in sRNATarBase and related literatures. (3) BSRD includes information on regulatory relationships between transcription factors (TF) and their target genes, which could provide insights into the combinatorial regulations of sRNAs and TF to their common targets. (4) BSRD has integrated expression data from NCBI GEO (Gene Expression Omnibus), which provides detailed evidences for sRNA expression profiling and re-annotation. (5) BSRD includes multiple new sRNA annotations from manually curated literature mining, including growth phase, Hfq binding, dual function and Rho-independent terminators. (6) BSRD harbors a novel RNA-Seq analysis platform, sRNADeep, that allows perform comprehensive sRNA expression profiling and differential expression analysis in large-scale transcriptome sequencing projects. With the aid of sRNADeep, users can (i) filter low-quality reads and adaptors from raw sequencing data, and (ii) align large amount of short reads to BSRD for identification of known sRNAs. (7) BSRD has implemented a Wikipedia-based community annotation function. (8) BSRD has a user-friendly interface, a flexible search option and a BLAST server for sequence homology searching. If you want to download all sRNA sequences in BSRD, you can access this link to get a quick download. Before getting started, you are suggested to read the full help document to get a quick guide on the database.
KBDOCK is a 3D database system that defines and spatially clusters protein binding sites for knowledge-based protein docking. KBDOCK extracts protein domain-domain interaction (DDI) and domain-peptide interaction (DPI) information from the PDB using the PFAM domain classification in order to analyse the spatial arrangements of DDIs and DPIs by Pfam family, and to propose structural templates for protein docking. Given a query Pfam domain, KBDOCK shows: - a non-redundant list of DDIs involving the query domain, grouped by their binding site.
- a Jmol view showing the DDIs placed in the coordinate frame of the query domain. You can choose to view one DDI at a time or a group of DDIs which share a similar binding site according to our binding site direction vector algorithm (see references).
- a consensus sequence alignment of the domains. Each sequence is annotated with core and rim interacting residues, and also the binding site centre residue.
Given a query Pfam domain, KBDOCK can also show DDIs involving different but structurally similar Pfam domains. For each Pfam family, KBDOCK stores a list of "Pfam neighbours" which is calculated using the Kpax structure alignment algorithm. Given two query domain structures, if full-homology (FH)† DDI templates exist in KBDOCK, then for each distinct interface, KBDOCK provides: - the best FH template (based on overall sequence identity) and the corresponding docking model.
- a Jmol view of the query domains superposed onto the FH template.
- a pairwise sequence alignment of the query domains and their corresponding template domain, showing the core, rim, and centre binding site residues.
If FH DDIs do not exist or if only one query domain structure is given, KBDOCK finds semi-homology (SH)† DDI templates. For each query domain and for each distinct binding site, it provides: - the best SH template (based on domain sequence identity).
- a Jmol view of the query domain superposed onto the SH template. A centre binding site residue is proposed.
- a pairwise sequence alignment of the query domain and its corresponding template domain, showing the core, rim, and centre binding site residues.
KBDOCK can also find DDIs involving structurally similar Pfam domains to the query domains using pre-calculated Pfam neighbour lists.
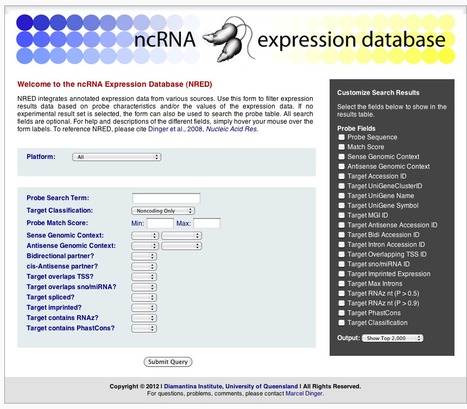
NRED integrates annotated expression data from various sources. Use this form to filter expression results data based on probe characteristics and/or the values of the expression data. If no experimental result set is selected, the form can also be used to search the probe table. All search fields are optional. For help and descriptions of the different fields, simply hover your mouse over the form labels. To reference NRED, please cite Dinger et al., 2008, Nucleic Acid Res.
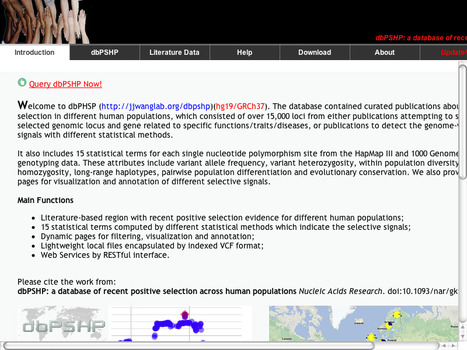
Welcome to dbPHSP (http://jjwanglab.org/dbpshp)(hg19/GRCh37). The database contained curated publications about positive selection in different human populations, which consisted of over 15,000 loci from either publications attempting to study positively selected genomic locus and gene related to specific functions/traits/diseases, or publications to detect the genome-wide selective signals with different statistical methods.
It also includes 15 statistical terms for each single nucleotide polymorphism site from the HapMap III and 1000 Genomes Project genotyping data. These attributes include variant allele frequency, variant heterozygosity, within population diversity, haplotype homozygosity, long-range haplotypes, pairwise population differentiation and evolutionary conservation. We also provided interactive pages for visualization and annotation of different selective signals.
The JASPAR CORE database contains a curated, non-redundant set of profiles, derived from published collections of experimentally defined transcription factor binding sites for eukaryotes. The prime difference to similar resources (TRANSFAC, etc) consist of the open data acess, non-redundancy and quality. When should it be used? When seeking models for specific factors or structural classes, or if experimental evidence is paramount
The iPfam database is a catalog of protein family interactions, including domain and ligand interactions, calculated from known structures. We take all known structures in the PDB, identify the protein domain(s) that are present in each structure and look for bonds between each domain and any other domains and/or small chemical ligands found in the structure. The bonds are automatically estimated based on both geometric and chemical properties of the two atoms involved.
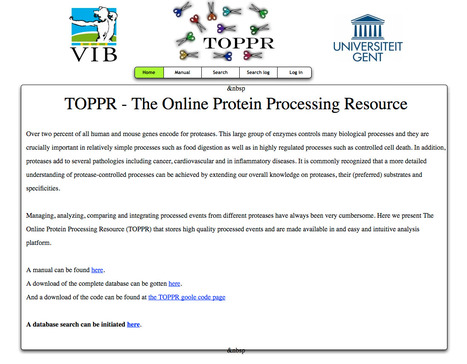
Over two percent of all human and mouse genes encode for proteases. This large group of enzymes controls many biological processes and they are crucially important in relatively simple processes such as food digestion as well as in highly regulated processes such as controlled cell death. In addition, proteases add to several pathologies including cancer, cardiovascular and in inflammatory diseases. It is commonly recognized that a more detailed understanding of protease-controlled processes can be achieved by extending our overall knowledge on proteases, their (preferred) substrates and specificities. Managing, analyzing, comparing and integrating processed events from different proteases have always been very cumbersome. Here we present The Online Protein Processing Resource (TOPPR) that stores high quality processed events and are made available in and easy and intuitive analysis platform. A manual can be found here. A download of the complete database can be gotten here. A database search can be initiated here.
InterDom is a database of putative interacting protein domains derived from multiple sources, ranging from domain fusions (Rosetta Stone), protein interactions (DIP and BIND), protein complexes (PDB), to scientific literature (MEDLINE).
Interdom focuses on providing supporting evidence for validating and annotating detected protein interactions and complexes based on putative protein domain interactions. InterDom enhances the quality of in silico derivations by adopting an integrative strategy, assigning higher confidence to domain interactions that are independently derived from different data sources and methods.
The current version of InterDom is 2.0, released July 31, 2007. Click here for the statistics data of InterDom v2.0
1. What kind of information can I find inside this database? SUPERTARGET is a database developed in the first place to collect informations about drug-target relations. It consist mainly of three different types of entities: - DRUGS
- PROTEINS
- SIDE-EFFECTS.
Beside this, informations about pathways and ontologies can be gained. Separate section is dedicated to a special soubgroup of the the targets - the cytochromes (CYPs) P450. All these entities are connected between each other through drug-protein, protein-protein and drug-side-effect relations and include rich annotation about the source, ID's, physical properties, references and much more. Proteins are retrieved from UniProt and are displayed with synonyms and organism information. Also information about target-target interactions and sequenc similarities between the targets are involved. Additional information as 3D-structures from PDB and EC-numbers are given if available. Drugs gained from SuperDrugwere mapped with BindingDB, integrated to SUPERTARGET and assigned metadata as ATC-codes (Anatomical Therapeutic Chemical), structure information and binding affinities. The side-effects contained in the database were fetched from SIDER and related to the drugs. 2. How was the data in the database created? The aim of SUPERTARGET is to offer an at most comprehensive datasets. Here for drug-target relations from different well-known databases as DrugBank,BindingDB and SuperCyp were integrated. To enhance the completeness of the dataset additional newly explored relations were incorporated, too. The new information was obtained in two steps: - Firstly, text-mining algorithms were applied to sort all PubMed listed papers by their relevance for drug-target relations.
- In a second step, the 7,000 papers with the highest rank were manually revised.
Protein-protein interaction data was obtained from ConsensusPathDB which integrates physical protein-protein interactions, metabolic and signaling reactions and gene regulatory interactions. Information on complex composition comes from Corum a protein complex database. Detailed overview about sources and relations between the entities presented by SUPERTARGET is summarized in following figure :
The BioMart Community Portal (www.biomart.org) is a community-driven effort to provide a unified interface to biomedical databases that are distributed worldwide. The portal provides access to numerous database projects supported by 30 scientific organizations. It includes over 800 different biological datasets spanning genomics, proteomics, model organisms, cancer data, ontology information and more. All resources available through the portal are independently administered and funded by their host organizations. The BioMart data federation technology provides a unified interface to all the available data. The latest version of the portal comes with many new databases that have been created by our ever-growing community. It also comes with better support and extensibility for data analysis and visualization tools. A new addition to our toolbox, the enrichment analysis tool is now accessible through graphical and web service interface. The BioMart community portal averages over one million requests per day. Building on this level of service and the wealth of information that has become available, the BioMart Community Portal has introduced a new, more scalable and cheaper alternative to the large data stores maintained by specialized organizations.
Via International Potato Center (CIP)
SuperTarget 2; AG Preissner; Charite Berlin
|
Listeria species are ubiquitous in the environment and often contaminate foods because they grow under conditions used for food preservation. Listeria monocytogenes, the human and animal pathogen, causes Listeriosis, an infection with a high mortality rate in risk groups such as immune-compromised individuals. Furthermore, L.monocytogenes is a model organism for the study of intracellular bacterial pathogens. The publication of its genome sequence and that of the non-pathogenic species Listeria innocua initiated numerous comparative studies and efforts to sequence all species comprising the genus. The Proteome database LEGER (http://leger2.gbf.de/cgi-bin/expLeger.pl) was developed to support functional genome analyses by combining information obtained by applying bioinformatics methods and from public databases to improve the original annotations. LEGER offers three unique key features: (i) it is the first comprehensive information system focusing on the functional assignment of genes and proteins; (ii) integrated visualization tools, KEGG pathway and Genome Viewer, alleviate the functional exploration of complex data; and (iii) LEGER presents results of systematic post-genome studies, thus facilitating analyses combining computational and experimental results. Moreover, LEGER provides an unpublished membrane proteome analysis of L.innocua and in total visualizes experimentally validated information about the subcellular localizations of 789 different listerial proteins.
mESAdb is a regularly updated database for the multivariate analysis of sequences and expression of microRNAs from multiple taxa. mESAdb is modular and has a user interface implemented in PHP and JavaScript and coupled with statistical analysis and visualization packages written for the R language. The database primarily comprises mature microRNA sequences and their target data, along with selected human, mouse and zebrafish expression datasets. mESAdb analysis modules allow a) mining of selected microRNA expression datasets for a list of microRNAs; and b) pair-wise multivariate analysis of expression datasets within and between taxa; and c) association of microRNA lists or microRNAs with a given motif with annotation databases, HUGE Navigator [1], KEGG [2], and GO [3]. The use of existing and customized R packages facilitates future addition of datasets and analysis tools. Furthermore, the ability to upload and analyze user-specified datasets makes mESAdb an interactive and expandable analysis tool for microRNA sequence and expression data.
KineticDB is the thoroughly curated database of protein folding kinetics which contains currently experiments on 87 unique proteins and about hundred of mutants. The main goal of KineticDB is to provide users with the diverse set of protein folding rates known from experiment. Currently the database contains kinetic data on single domain proteins, separate protein domains and short polypeptides without S-S bonds in the native structure.
We hope that the database will be useful for you. Help us make the database as wide and up-to-date as possible - send us references containing new kinetic data! We will be grateful for any contribution to the database from the community: for bug reports, for new kinetic data and for any questions
How to cite:
Bogatyreva N.S., Osypov A.A., Ivankov D.N. (2009). KineticDB: a database of protein folding kinetics. NAR, 37:D342-D346.
With the completion of the sequencing of the genomes of human and other organisms, attention has focused on the characterization and function of proteins, the products of genes. The availability of sequence data and the growing impact of structural biology on biomedical research have prompted scientific groups from several countries to undertake projects in the emerging field of structural genomics. The objective is to make these structures widely available for clinical and basic studies that will expand the knowledge of the role of proteins both in normal biological processes and in disease. The National Institute of General Medical Sciences (NIGMS) played a major role in the early planning for structural genomics and in 1999 organized a national program, the Protein Structure Initiative. The PSI program officially concluded in 2015, having determined nearly 7000 protein structures, developed ~450 new or improved technologies, and wrote over 2200 publications. The impact of this work enabled the greater biological and biomedical communities.
The Fungal Program scales up sequencing and analysis of fungal genomes to explore the diversity of fungi important for energy and the environment, and to promote functional studies on a system level. Encoded in the genomes of the organisms of the kingdom Fungi are biological processes with high relevance to the Department of Energy missions in bioenergy production, carbon cycling and biogeochemistry. Combining new sequencing technologies and comparative genomics analysis, we work on large and complex sequencing projects such as surveying the broad phylogenetic and ecological diversity of fungi, and capturing genomic variation in natural populations and engineered strains. This approach allows us to build a foundation for translating the genomic potential of fungi into practical applications.
Optimal global pairwise alignments from about 270,000 ribosomal RNA (rRNA) internal transcribed spacer 2 (ITS2) sequences - all against all - have been generated in order to model ITS2 secondary structures based on sequences with known structures. Via 60,000 known ITS2 sequences that fit a common core of the ITS2 secondary structure described for the eukaryotes (Schultz et al. 2005), homology based modeling (Wolf et al. 2005) and reannotation procedures revealed in addition more than 150,000 homologous structures that could not be predicted by standard RNA folding programs.
iPAVS provides a collection of highly-structured manually curated human pathway data, it also integrates biological pathway information from several public databases and provides several tools to manipulate,filter, browse, search, analyze, visualize and compare the integrated pathway resources.
iPAVS has implemented advanced functionalities to perform complex pathway analysis tasks and visualize the results. But, at the same time extensive efforts has been put to design the user interface simple and intuitive enough for biologists to use IPAVS functionalities with ease. Main intention behind launching iPAVS is to enable biologists to do systems biology and employ systems methodologies in their day to day research and produce testable hypothesis, without having to engage themselves in difficult, resource demanding and time consuming data integration and management tasks.
Publication:
Title: IPAVS: Integrated Pathway Resources, Analysis and Visualization System
Author(s): Pradeep Kumar Sreenivasaiah, Shilpa Rani, Joseph Cayetano, Novino Arul, and Do Han Kim
Source: Nucleic Acids Research Published online: December 2, 2011 : gkr1208v1-gkr1208.
DOI: 10.1093/nar/gkr1208
InvFEST aims to become a reference site to share information and collaborate towards the complete characterization of human polymorphic inversions. It is a data-warehouse implementation that integrates several data of interest related to inversions with an online analytical processing engine (OLAP) to gather information and compute a report of each inversion. The InvFEST database stores and merges inversion predictions from healthy individuals into a non-redundant dataset by overlapping the position of the breakpoints of each prediction and taking into account the resolution of each study. Most predictions come from mapping information of paired-end sequences (PEM) obtained by different studies of the literature, which in some cases have been reanalyzed by GRIAL, a program specifically designed to detect inversions from PEM data. Moreover, it stores information of validations and genotyping assays, frequency in different populations, association with genes and segmental duplications, and the evolutionary history of the inversions. The database will keep on updating information by incorporating new predictions, validations, genotyping data, and any other information, either extracted from peer reviewed research studies or generated in our lab. We always welcome your suggestions and comments. The InvFEST database is an outcome of the INVFEST project, supported by the European Research Council (ERC) Starting Grant 243212 under the European Union Seventh Research Framework Programme (FP7).
CTCF is a highly conserved transcriptional regulator protein that performs diverse functions such as regulating gene expression and organizing the 3D structure of the genome. Here, we describe recent updates to a database of CTCF-binding sites, CTCFBSDB (http://insulatordb.uthsc.edu/), which now contains almost 15 million CTCF-binding sequences in 10 species. Since the original publication of the database, studies of the 3D structure of the genome, such as those provided by Hi-C experiments, have suggested that CTCF plays an important role in mediating intra- and inter-chromosomal interactions. To reflect this important progress, we have integrated CTCF-binding sites with genomic topological domains defined using Hi-C data. Additionally, the updated database includes new features enabled by new CTCF-binding site data, including binding site occupancy and the ability to visualize overlapping CTCF-binding sites determined in separate experiments.
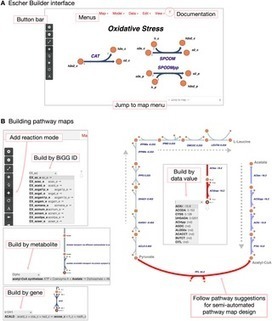
Escher is a web application for visualizing data on biological pathways. Three key features make Escher a uniquely effective tool for pathway visualization. First, users can rapidly design new pathway maps. Escher provides pathway suggestions based on user data and genome-scale models, so users can draw pathways in a semi-automated way. Second, users can visualize data related to genes or proteins on the associated reactions and pathways, using rules that define which enzymes catalyze each reaction. Thus, users can identify trends in common genomic data types (e.g. RNA-Seq, proteomics, ChIP)—in conjunction with metabolite- and reaction-oriented data types (e.g. metabolomics, fluxomics). Third, Escher harnesses the strengths of web technologies (SVG, D3, developer tools) so that visualizations can be rapidly adapted, extended, shared, and embedded. This paper provides examples of each of these features and explains how the development approach used for Escher can be used to guide the development of future visualization tools.
Via Biswapriya Biswavas Misra
|



 Your new post is loading...
Your new post is loading...


















Each person’s experience with acid will be different. Indeed, each trip may be different. One might be very light, but others can take a frightening and overwhelming turn.
Symptoms may begin to show 20 to 90 minutesTrusted Source after taking a dose. The main episode can last several hours.
Acid is a long-acting drug. It stays in the body 6 to 15 hoursTrusted Source. Most acid trips won’t last more than 9 hours.
The trip
During this period of “tripping” or active effects, you may begin to experience sensationalized perceptions of what’s happening around you. This can include “seeing” color or “tasting” sounds. Stationary items, like furniture, may begin to “move” or swell or shrink before your eyes.
Coming down
Coming down from the trip will feel like you’re gradually returning to Earth. Signs may begin to lessen in intensity. You may feel tired after getting through the hours-long trip and want to sleep.
Afterglow
An “afterglow” is possible for several hours after the end of the trip, too. This may feel like everything is “lighter” or “brighter” than before the trip. You may also have moments of flashbacks for several hours, even days, after the acid trip is over.
Microdosing
A microdose is a small dose of a psychedelic drug like LSD, often one-tenth of a normal dose. It’s sometimes used to help treat symptoms of anxiety and depression, but it’s not meant to completely encompass your day. However, little is known about the long-term effects of this practice.
Visit Howhighcenter.com for some Psychedelic Products..
https://howhighcenter.com/shop/chocolate-bars/magic-chocolate-caps/
https://howhighcenter.com/shop/chocolate-bars/wavy-bar-chocolate-mushroom/
https://howhighcenter.com/shop/chocolate-bars/magic-chocolate-caps/
https://howhighcenter.com/shop/chocolate-bars/trippy-flip-milk-chocolate-bar/
https://howhighcenter.com/shop/chocolate-bars/one-up-mushroom-bar-3-5g-shrooms-chocolate-bar/
https://howhighcenter.com/shop/chocolate-bars/rick-and-morty-trippy-flip/
https://howhighcenter.com/shop/shrooms/columbia-cap/
https://howhighcenter.com/shop/chocolate-bars/trippy-flip-milk-chocolate-bar/
https://howhighcenter.com/shop/chocolate-bars/charming-bars/
https://howhighcenter.com/shop/chocolate-bars/one-up-mushroom/
https://howhighcenter.com/shop/chocolate-bars/polkadot-magic-muchroom-belgian-bar/
https://howhighcenter.com/shop/shrooms/blue-meanies-mushrooms/
https://howhighcenter.com/shop/chocolate-bars/rick-and-morty-trippy-flip/
https://howhighcenter.com/shop/chocolate-bars/blue-meanies-mushroom/
https://howhighcenter.com/shop/chocolate-bars/charming-bars/
https://howhighcenter.com/shop/chocolate-bars/dark-chocolate-bars/
https://howhighcenter.com/shop/chocolate-bars/magic-chocolate-caps/
https://howhighcenter.com/shop/chocolate-bars/one-up-mushroom/
https://howhighcenter.com/shop/chocolate-bars/one-up-mushroom-bar-3-5g-shrooms-chocolate-bar/
https://howhighcenter.com/shop/chocolate-bars/open-minded-chocolate-bars/
https://howhighcenter.com/shop/chocolate-bars/polkadot-magic-muchroom-belgian-bar/
https://howhighcenter.com/shop/chocolate-bars/rick-and-morty-trippy-flip/
https://howhighcenter.com/shop/chocolate-bars/space-bars-dark-chocolate/
https://howhighcenter.com/shop/chocolate-bars/trippy-flip-milk-chocolate-bar/
https://howhighcenter.com/shop/chocolate-bars/trippy-treats-magic-shrooms-chocolate-bars/
https://howhighcenter.com/shop/chocolate-bars/wavy-bar-chocolate-mushroom/
https://howhighcenter.com/shop/shrooms/albino-penis-envy-mushrooms/
https://howhighcenter.com/shop/shrooms/amanita-muscaria/
https://howhighcenter.com/shop/shrooms/amazon-cubensis/
https://howhighcenter.com/shop/shrooms/blue-meanies-mushrooms/
https://howhighcenter.com/shop/shrooms/brazilian-cubensis/
https://howhighcenter.com/shop/shrooms/columbia-cap/
https://howhighcenter.com/shop/shrooms/dancing-tiger/
https://howhighcenter.com/shop/shrooms/golden-caps/
https://howhighcenter.com/shop/shrooms/golden-teachers/
https://howhighcenter.com/shop/shrooms/hawaiian-caps/
https://howhighcenter.com/shop/shrooms/p-e-shrooms/
https://howhighcenter.com/shop/shrooms/psilocybe-azurescens-flying-saucer-mushrooms/
https://howhighcenter.com/shop/lsd/25i-nbome-blotter/
https://howhighcenter.com/shop/lsd/liquid-lsd/
https://howhighcenter.com/shop/lsd/liquid-lsd-acid-100/
https://howhighcenter.com/shop/lsd/liquid-lsd-acid-200/
https://howhighcenter.com/shop/lsd/lsd-blotter-200-%c2%b5g/
https://howhighcenter.com/shop/lsd/lsd-blotter-250-%c2%b5g/
https://howhighcenter.com/shop/lsd/lsd-sheets/
https://howhighcenter.com/shop/dmt/5-meo-dmt/
https://howhighcenter.com/shop/dmt/crimson-palms-hotel/
https://howhighcenter.com/shop/dmt/dmt-vape/
https://howhighcenter.com/shop/dmt/dmt-vape-top-diy-refill-syringe/
https://howhighcenter.com/shop/dmt/mdma-powder/
https://howhighcenter.com/shop/dmt/raw-dmt-powder-1g/
https://howhighcenter.com/shop/dmt/somad-dmt-vape-top/