Your new post is loading...
Scientists in Barcelona have today launched GENIGMA, a videogame that enlists players to solve puzzles while generating real-world scientific data that can detect alterations in genomic sequences and ultimately advance breast cancer research. The game, out today on iOS and Android and available in English, Spanish, Catalan and Italian, is the result of a two-and-a-half-year long citizen science project developed by a team of researchers at the Centre for Genomic Regulation (CRG), the Centro Nacional de Análisis Genómico (CNAG-CRG) and game professionals. The game was created to boost worldwide research efforts that depend on cancer cell lines, a critical resource used by scientists to study cancer and test new drugs to treat the disease. One of the limitations of cancer cell lines are a lack of high-resolution genome reference maps, which are necessary to help researchers interpret their scientific results, for example pinpointing the location of genes of therapeutic interest or potential mutation sites. "Cell lines are responsible for the discovery of vaccines, chemotherapies for cancer or IVF for infertility. This makes them a pillar of modern biology," explains ICREA Research Professor Marc A. Marti-Renom, with dual affiliation at the CRG and CNAG-CRG and whose research underpins GENIGMA. "However, the lack of genome reference maps limits current scientific progress. It's like asking people to navigate modern cities using maps from the past. With the help of other people, we can update these maps, which will allow us to make fast progress in breast cancer research." Professor Marti-Renom's research group has developed methods to create genomic reference maps by visualising the genome in three-dimensional space. However, this requires significant time and resources to train artificial intelligence, as well as vast computational power. The researchers launched GENIGMA because they believe that data generated by players could be a more effective method of updating the reference maps compared to using AI alone. The 'herd intelligence' of players can also provide creative solutions in ways that AI might not be able to. To play GENIGMA, players have to solve a puzzle involving a string of blocks of different colours and shapes. Each string represents a genetic sequence in the cancer cell line, and how players organise the blocks is a potential solution to the location of genes. Players have to reorganise the blocks so that they attain the highest-score possible. The higher the number of players and high scores, the higher likelihood that researchers have found the correct sequence for this particular location in the reference map. "Anyone with a smartphone from anywhere in the world can download GENIGMA for free and make a direct contribution to research, lending their logic and dexterity to the service of science," says Elisabetta Broglio, citizen science facilitator at the CRG. "GENIGMA will analyze the solutions provided by the players as a collective and not as individuals, and will take advantage of creative solutions impossible to find with deterministic algorithms."
Broad Institute of MIT and Harvard is teaming up with Google Genomics to explore how to break down major technical barriers that increasingly hinder biomedical research by addressing the need for computing infrastructure to store and process enormous datasets, and by creating tools to analyze such data and unravel long-standing mysteries about human health.
As a first step, Broad Institute’s Genome Analysis Toolkit, or GATK, will be offered as a service on the Google Cloud Platform, as part of Google Genomics. The goal is to enable any genomic researcher to upload, store, and analyze data in a cloud-based environment that combines the Broad Institute’s best-in-class genomic analysis tools with the scale and computing power of Google.
GATK is a software package developed at the Broad Institute to analyze high-throughput genomic sequencing data. GATK offers a wide variety of analysis tools, with a primary focus on genetic variant discovery and genotyping as well as a strong emphasis on data quality assurance. Its robust architecture, powerful processing engine, and high-performance computing features make it capable of taking on projects of any size.
GATK is already available for download at no cost to academic and non-profit users. In addition, business users can license GATK from the Broad. To date, more than 20,000 users have processed genomic data using GATK.
The Google Genomics service will provide researchers with a powerful, additional way to use GATK. Researchers will be able to upload genetic data and run GATK-powered analyses on Google Cloud Platform, and may use GATK to analyze genetic data already available for research via Google Genomics. GATK as a service will make best-practice genomic analysis readily available to researchers who don’t have access to the dedicated compute infrastructure and engineering teams required for analyzing genomic data at scale. An initial alpha release of the GATK service will be made available to a limited set of users.
“Large-scale genomic information is accelerating scientific progress in cancer, diabetes, psychiatric disorders, and many other diseases,” said Eric Lander, President and Director of Broad Institute. “Storing, analyzing, and managing these data is becoming a critical challenge for biomedical researchers. We are excited to work with Google’s talented and experienced engineers to develop ways to empower researchers around the world by making it easier to access and use genomic information.”
New insights into the behavior of photosynthetic proteins from atomic simulations could hasten the development of artificial light-gathering machines.
The protein complex known as photosystem II splits water molecules to release oxygen using sunlight and relatively simple biological building blocks. Although water can also be split artificially using an electrical voltage and a precious metal catalyst, researchers continue to strive to mimic the efficient natural process. So far, however, these efforts have been hampered by an incomplete understanding of the water oxidation mechanism of photosystem II. Shinichiro Nakamura from the RIKEN Innovation Center and colleagues have now used simulations to reveal the hidden pathways of water molecules inside photosystem II.
At the heart of photosystem II is a cluster of manganese, calcium and oxygen atoms, known as the oxygen-evolving complex (OEC), that catalyzes the water-splitting reaction. Recent high-resolution x-ray crystallography studies have revealed the precise positions of the atoms in the OEC and of the protein residues that contact the site. While this information has yielded important structural clues into photosynthetic water oxidation, the movements of water, oxygen and protons within the protein complex are still the subject of much speculation.
To resolve this problem, researchers have turned to molecular dynamics (MD) simulation, a technique that models the time-dependent behavior of biomolecules using thermodynamics and the physics of motion. While previous MD simulations of photosystem II have involved the use of approximate models that focus only on protein monomers or the main OEC components, Nakamura's team took a different approach. "Our hypothesis was that we cannot understand the mechanism of oxygen evolution just by looking at the manganese-based reaction center," he says. "Therefore, we carried out a total MD simulation, without any truncation of the protein or simplification."
In their simulation, the team embedded an exact model of photosystem II inside a thylakoid—a lipid and fatty-acid membrane-bound compartment found in the chloroplasts of plant cells. After initial computations confirmed the reliability of their model, the researchers performed a rigorous MD simulation of the protein—membrane system in the presence of more than 300,000 water molecules (Fig. 1). "The results indicated that water, oxygen and protons move through photosystem II not randomly but via distinct pathways that are not obviously visible," says Nakamura.
The pathways revealed by the simulations are delicately coupled to the dynamic motions of the photosystem II protein residues. While such intricate activity is currently impossible to reproduce artificially, the researchers suspect that combining quantum-chemical calculations with MD simulations could help to unlock the mysterious principles behind the highly efficient oxygen-evolution reactions of this remarkable biological factory.
Turning vast amounts of genomic data into meaningful information about the cell is the great challenge of bioinformatics, with major implications for human biology and medicine. Researchers at the University of California, San Diego School of Medicine and colleagues have proposed a new method that creates a computational model of the cell from large networks of gene and protein interactions, discovering how genes and proteins connect to form higher-level cellular machinery.
"Our method creates ontology, or a specification of all the major players in the cell and the relationships between them," said first author Janusz Dutkowski, PhD, postdoctoral researcher in the UC San Diego Department of Medicine. It uses knowledge about how genes and proteins interact with each other and automatically organizes this information to form a comprehensive catalog of gene functions, cellular components, and processes.
"What's new about our ontology is that it is created automatically from large datasets. In this way, we see not only what is already known, but also potentially new biological components and processes – the bases for new hypotheses," said Dutkowski.
Originally devised by philosophers attempting to explain the nature of existence, ontologies are now broadly used to encapsulate everything known about a subject in a hierarchy of terms and relationships. Intelligent information systems, such as iPhone's Siri, are built on ontologies to enable reasoning about the real world. Ontologies are also used by scientists to structure knowledge about subjects like taxonomy, anatomy and development, bioactive compounds, disease and clinical diagnosis.
|
Synthetic biologists have created software that automates the design of DNA circuits for living cells. The aim is to help people who are not skilled biologists to quickly design working biological systems, says synthetic biologist Christopher Voigt at the Massachusetts Institute of Technology in Cambridge, who led the work. “This is the first example where we’ve literally created a programming language for cells,” he says. In the new software — called Cello — a user first specifies the kind of cell they are using and what they want it to do: for example, sense metabolic conditions in the gut and produce a drug in response. They type in commands to explain how these inputs and outputs should be logically connected, using a computing language called Verilog that electrical engineers have long relied on to design silicon circuits. Finally, Cello translates this information to design a DNA sequence that, when put into a cell, will execute the demands. Voigt says his team is writing user interfaces that would allow biologists to write a single program and be returned different DNA sequences for different organisms. Anyone can access Cello through a Web-based interface, or by downloading its open-source code from the online repository GitHub. ”This paper solves the problem of the automated design, construction and testing of logic circuits in living cells,” says bioengineer Herbert Sauro at the University of Washington in Seattle, who was not involved in the study. The work is published in Science.1
Some of the world’s most brilliant minds are working as data scientists at places like Google, Facebook, and Twitter—analyzing the enormous troves of online information generated by these tech giants—and for hacker and entrepreneur Jeremy Howard, that’s a bit depressing. Howard, a data scientist himself, spent a few years as the president of the Kaggle, a kind of online data scientist community that sought to feed the growing thirst for information analysis. He came to realize that while many of Kaggle’s online data analysis competitions helped scientists make new breakthroughs, the potential of these new techniques wasn’t being fully realized. “Data science is a very sexy job at the moment,” he says. “But when I look at what a lot of data scientists are actually doing, the vast majority of work out there is on product recommendations and advertising technology and so forth.”
So, after leaving Kaggle last year, Howard decided he would find a better use for data science. Eventually, he settled on medicine. And he even did a kind of end run around the data scientists, leveraging not so much the power of the human brain but the rapidly evolving talents of artificial brains. His new company is called Enlitic, and it wants to use state-of-the-art machine learning algorithms—what’s known as “deep learning”—to diagnosis illness and disease.
Publicly revealed for the first time today, the project is only just getting off the ground—“the big opportunities are going to take years to develop,” Howard says—but it’s yet another step forward for deep learning, a form of artificial intelligence that more closely mimics the way our brains work. Facebook is exploring deep learning as a way of recognizing faces in photos. Google uses it for image tagging and voice recognition. Microsoft does real-time translation in Skype. And the list goes on.
But Howard hopes to use deep learning for something more meaningful. His basic idea is to create a system akin to the Star Trek Tricorder, though perhaps not as portable. Enlitic will gather data about a particular patient—from medical images to lab test results to doctors’ notes—and its deep learning algorithms will analyze this data in an effort to reach a diagnosis and suggest treatments. The point, Howard says, isn’t to replace doctors, but to give them the tools they need to work more effectively. With this in mind, the company will share its algorithms with clinics, hospitals, and other medical outfits, hoping they can help refine its techniques. Howard says that the health care industry has been slow to pick-up on the deep-learning trend because it was rather expensive to build the computing clusters needed to run deep learning algorithms. But that’s changing.
The real challenge, Howard says, isn’t writing algorithms but getting enough data to train those algorithms. He says Enlitic is working with a number of organizations that specialize in gathering anonymized medical data for this type of research, but he declines to reveal the names of the organizations he’s working with. And while he’s tight-lipped about the company’s technique now, he says that much of the work the company does will eventually be published in research papers.
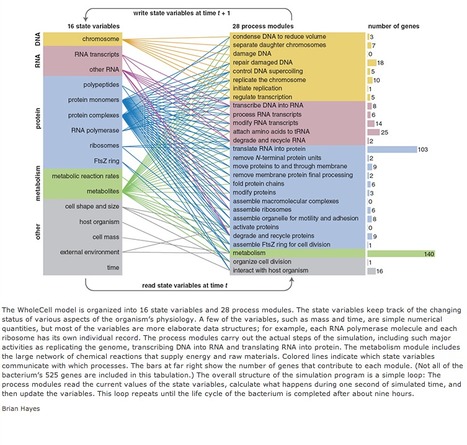
Almost 30 years ago, Harold J. Morowitz, who was then at Yale, set forth a bold plan for molecular biology. He outlined a campaign to study one of the smallest single-celled organisms, a bacterium of the genus Mycoplasma. The first step would be to decipher its complete genetic sequence, which in turn would reveal the amino acid sequences of all the proteins in the cell. In the 1980s reading an entire genome was not the routine task it is today, but Morowitz argued that the analysis should be possible if the genome was small enough. He calculated the information content of mycoplasma DNA to be about 160,000 bits, then added: Alternatively, this much DNA will code for about 600 proteins—which suggests that the logic of life can be written in 600 steps. Completely understanding the operations of a prokaryotic cell is a visualizable concept, one that is within the range of the possible. There was one more intriguing element to Morowitz’s plan: At 600 steps, a computer model is feasible, and every experiment that can be carried out in the laboratory can also be carried out on the computer. The extent to which these match measures the completeness of the paradigm of molecular biology. Looking back on these proposals from the modern era of industrial-scale genomics and proteomics, there’s no doubt that Morowitz was right about the feasibility of collecting sequence data. On the other hand, the challenges of writing down “the logic of life” in 600 steps and “completely understanding” a living cell still look fairly daunting. And what about the computer program that would simulate a living cell well enough to match experiments carried out on real organisms? As it happens, a computer program with exactly that goal was published last summer by Markus W. Covert of Stanford University and eight coworkers. The program, called the WholeCell simulation, describes the full life cycle of Mycoplasma genitalium, a bacterium from the genus that Morowitz had suggested. Included in the model are all the major processes of life: transcription of DNA into RNA, translation of RNA into protein, metabolism of nutrients to produce energy and structural constituents, replication of the genome, and ultimately reproduction by cell fission. The outputs of the simulation do seem to match experimental results. So the question has to be faced: Are we on the threshold of “completing” molecular biology?
|



 Your new post is loading...
Your new post is loading...