 Your new post is loading...

|
Scooped by
Juan Lama
|
RetroVirox offers a menu of cell-based antiviral services to evaluate experimental therapies and vaccines against coronaviruses, including SARS-CoV-2. The company offers in vitro testing with SARS-CoV-2 pseudovirions and with live SARS-CoV-2 viruses to evaluate entry inhibitors, neutralizing antibodies, and antivirals against the novel coronavirus causative agent of COVID-19. Multiple viral strains are available for testing, including the most recent Omicron variants of concern (XBB.1.5, XBB.1.16, XBB.2.3.2, EG.5.1 and JN.1). Pseudoviruses coated with the viral spike (S) protein of SARS-CoV-2 are also used to recapitulate the mode of entry of the novel coronavirus. Over 50 spike mutant and variants are available as pseudoviruses. These assays can be used for the following purposes: - To determine the neutralizing activity of therapeutic antibodies and antisera
- To test experimental COVID-19 vaccines using antisera from inoculated animals or humans
- To evaluate small-molecule and other entry inhibitors targeting the S viral protein, the ACE-2 viral receptor, or host proteases and other targets involved in SARS-CoV-2 viral entry
Assays with live replicating SARS-CoV-2, and milder forms of seasonal human coronaviruses (hCoV-OC43 and 229E) allow for the evaluation of inhibitors at all stages of the coronavirus life cycle. Additional Information about the coronavirus assays and many other cell-based antiviral assays offered at RetroVirox is available here. Request additional information by email at info@retrovirox.com

|
Scooped by
Juan Lama
|
Respiratory virus infections in humans cause a broad-spectrum of diseases that result in substantial morbidity and mortality annually worldwide. To reduce the global burden of respiratory viral diseases, preventative and therapeutic interventions that are accessible and effective are urgently needed, especially in countries that are disproportionately affected. Repurposing generic medicine has the potential to bring new treatments for infectious diseases to patients efficiently and equitably. In this study, we found that intranasal delivery of neomycin, a generic aminoglycoside antibiotic, induces the expression of interferon-stimulated genes (ISGs) in the nasal mucosa that is independent of the commensal microbiota. Prophylactic or therapeutic administration of neomycin provided significant protection against upper respiratory infection and lethal disease in a mouse model of COVID-19. Furthermore, neomycin treatment protected Mx1 congenic mice from upper and lower respiratory infections with a highly virulent strain of influenza A virus. In Syrian hamsters, neomycin treatment potently mitigated contact transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In healthy humans, intranasal application of neomycin-containing Neosporin ointment was well tolerated and effective at inducing ISG expression in the nose in a subset of participants. These findings suggest that neomycin has the potential to be harnessed as a host-directed antiviral strategy for the prevention and treatment of respiratory viral infections. Published in PNAS (April 22, 2024): https://doi.org/10.1073/pnas.2319566121

|
Scooped by
Juan Lama
|
Recommendations for Worker Protection and Use of Personal Protective Equipment (PPE) to Reduce Exposure to Highly Pathogenic Avian Influenza A H5 Viruses - CDC Information for workers Any person working with or exposed to animals such as poultry and livestock farmers and workers, backyard bird flock owners, veterinarians and veterinary staff, and responders should take steps to reduce the risk of infection with avian influenza A viruses associated with severe disease when working with animals or materials potentially infected or confirmed to be infected with these viruses. Avoid unprotected direct or close physical contact with: - Sick birds, livestock, or other animals
- Carcasses of birds, livestock, or other animals
- Feces or litter
- Raw milk
- Surfaces and water (e.g., ponds, waterers, buckets, pans, troughs) that might be contaminated with animal excretions.
If you must work with or enter any not yet disinfected buildings where these materials or sick or dead animals potentially infected or confirmed to be infected are or were present, wear appropriate personal protective equipment (PPE) in addition to the PPE you might be using for your normal duties (e.g., waterproof apron, hearing protection, etc.). Appropriate PPE depends on the hazards present and a site-specific risk assessment. If you have questions on the type of PPE to use or how to fit it properly, ask your supervisor. Recommended PPE to protect against novel influenza A viruses includes: - Disposable or non-disposable fluid-resistant [i] coveralls, and depending on task(s), add disposable or non-disposable waterproof apron
- Any NIOSH Approved® particulate respirator (e.g., N95® or greater filtering facepiece respirator, elastomeric half mask respirator with a minimum of N95 filters)
- Properly-fitted unvented or indirectly vented safety goggles [ii] or a faceshield if there is risk of liquid splashing onto the respirator
- Rubber boots or rubber boot covers with sealed seams that can be sanitized or disposable boot covers for tasks taking a short amount of time
- Disposable or non-disposable head cover or hair cover
- Disposable or non-disposable gloves [iii]

|
Scooped by
Juan Lama
|
Researchers at UC Riverside have developed a new vaccine approach using RNA that is effective against any strain of a virus and can be used safely even by babies or the immunocompromised. Every year, researchers try to predict the four influenza strains that are most likely to be prevalent during the upcoming flu season. And every year, people line up to get their updated vaccine, hoping the researchers formulated the shot correctly. The same is true of COVID vaccines, which have been reformulated to target sub-variants of the most prevalent strains circulating in the U.S. This new strategy would eliminate the need to create all these different shots, because it targets a part of the viral genome that is common to all strains of a virus. The vaccine, how it works, and a demonstration of its efficacy in mice is described in a paper published today in the Proceedings of the National Academy of Sciences. “What I want to emphasize about this vaccine strategy is that it is broad,” said UCR virologist and paper author Rong Hai. “It is broadly applicable to any number of viruses, broadly effective against any variant of a virus, and safe for a broad spectrum of people. This could be the universal vaccine that we have been looking for. Traditionally, vaccines contain either a dead or modified, live version of a virus. The body’s immune system recognizes a protein in the virus and mounts an immune response. This response produces T-cells that attack the virus and stop it from spreading. It also produces “memory” B-cells that train your immune system to protect you from future attacks. The new vaccine also uses a live, modified version of a virus. However, it does not rely on the vaccinated body having this traditional immune response or immune active proteins — which is the reason it can be used by babies whose immune systems are underdeveloped, or people suffering from a disease that overtaxes their immune system. Instead, this relies on small, silencing RNA molecules. Mechanism and Efficacy of RNA-Based Vaccine “A host — a person, a mouse, anyone infected— will produce small interfering RNAs as an immune response to viral infection. These RNAi then knock down the virus,” said Shouwei Ding, distinguished professor of microbiology at UCR, and lead paper author. The reason viruses successfully cause disease is because they produce proteins that block a host’s RNAi response. “If we make a mutant virus that cannot produce the protein to suppress our RNAi, we can weaken the virus. It can replicate to some level, but then loses the battle to the host RNAi response,” Ding said. “A virus weakened in this way can be used as a vaccine for boosting our RNAi immune system.” When the researchers tested this strategy with a mouse virus called Nodamura, they did it with mutant mice lacking T and B cells. With one vaccine injection, they found the mice were protected from a lethal dose of the unmodified virus for at least 90 days. Note that some studies show nine mouse days are roughly equivalent to one human year. There are few vaccines suitable for use in babies younger than six months old. However, even newborn mice produce small RNAi molecules, which is why the vaccine protected them as well. UC Riverside has now been issued a US patent on this RNAi vaccine technology. In 2013, the same research team published a paper showing that flu infections also induce us to produce RNAi molecules. “That’s why our next step is to use this same concept to generate a flu vaccine, so infants can be protected. If we are successful, they’ll no longer have to depend on their mothers’ antibodies,” Ding said. Their flu vaccine will also likely be delivered in the form of a spray, as many people have an aversion to needles. “Respiratory infections move through the nose, so a spray might be an easier delivery system,” Hai said. Additionally, the researchers say there is little chance of a virus mutating to avoid this vaccination strategy. “Viruses may mutate in regions not targeted by traditional vaccines. However, we are targeting their whole genome with thousands of small RNAs. They cannot escape this,” Hai said. Ultimately, the researchers believe they can ‘cut and paste’ this strategy to make a one-and-done vaccine for any number of viruses. “There are several well-known human pathogens; dengue, SARS, COVID. They all have similar viral functions,” Ding said. “This should be applicable to these viruses in an easy transfer of knowledge.” Publised in PNAS (April 17, 2024): https://doi.org/10.1073/pnas.2321170121

|
Scooped by
Juan Lama
|
Over four years have passed since the beginning of the COVID-19 pandemic. The scientific response has been rapid and effective, with many therapeutic monoclonal antibodies and small molecules developed for clinical use. However, given the ability for viruses to become resistant to antivirals, it is perhaps no surprise that the field has identified resistance to nearly all of these compounds. Here, we provide a comprehensive review of the resistance profile for each of these therapeutics. We hope that this resource provides an atlas for mutations to be aware of for each agent, particularly as a springboard for considerations for the next generation of antivirals. Finally, we discuss the outlook and thoughts for moving forward in how we continue to manage this, and the next, pandemic. Published (April 18, 2024): https://doi.org/10.1016/j.chembiol.2024.03.008

|
Scooped by
Juan Lama
|
As the virus continues its spread to new species, the World Health Organization fears it is moving closer to people. The risk of bird flu jumping into humans is of “enormous concern,” the World Health Organization has warned as the virus continues its spread into new species. Since 2020, an outbreak of the H5N1 strain has killed tens of millions of birds worldwide, along with thousands of mammals, including sea lions, elephant seals and even one polar bear. More recently, the virus emerged in 16 herds of cattle in Texas – a development which has surprised experts, as it was previously believed the animals were not susceptible to infection, and raised concerns that H5N1 could eventually spill over into humans. “This remains, I think, an enormous concern,” Dr Jeremy Farrar, the WHO’s chief scientist, told reporters on Thursday in Geneva. “When you come into the mammalian population, then you’re getting closer to humans … this virus is just looking for new, novel hosts.” He warned that, by circulating in new mammals, the virus improves its chances of further evolving and developing “the ability to infect humans and then critically the ability to go from human to human”. In the recent cattle outbreak in Texas, one person was diagnosed with bird flu following close contact with dairy cows presumed to have been infected. Over the past 20 years, hundreds of others have caught the virus following exposure to an infected animal – yet there has been no evidence of human-to-human transmission in these cases. Yet the mortality rate for humans is “extraordinarily high,” Dr Farrar warned. From 2003 to 2024, 463 deaths and 889 cases were reported from 23 countries, according to the WHO – a case fatality rate of 52 per cent. The Texas infection is the second case of bird flu in the US and appears to have been the first infection of the H5N1 strain through contact with an infected mammal, said the WHO. Dr Farrar said the ongoing bird flu outbreak had become “a global zoonotic animal pandemic” and called for increased monitoring, adding that it was “very important” to understand how many human infections are occuring “because that’s where adaptation [of the virus] will happen”. He added: “It’s a tragic thing to say, but if I get infected with H5N1 and I die, that’s the end of it. If I go around the community and I spread it to somebody else then you start the cycle.” He said efforts were being made to develop vaccines and therapeutics for the strain and stressed the importance of international health authorities having the capacity to diagnose the virus. This would ensure that the world would be “in a position to immediately respond” if the strain “did come across to humans, with human-to-human transmission”, said Dr Farrar.

|
Scooped by
Juan Lama
|
The results could pave the way for new therapeutic strategies using existing drugs to combat the growing AD crisis. Summary: Common HIV drugs could reduce the incidence of Alzheimer’s disease (AD). Utilizing anonymized prescription data from over 225,000 individuals, the study found that HIV-positive patients taking reverse transcriptase (RT) inhibitors showed a significantly lower rate of AD compared to the general population. This discovery builds on previous findings that Alzheimer’s-linked genes might be recombined by enzymes similar to those targeted by HIV treatments. The results could pave the way for new therapeutic strategies using existing drugs to combat the growing AD crisis. Key Facts: - The study analyzed prescription data from 225,000 individuals, revealing that HIV-positive patients over 60 taking RT inhibitors had fewer Alzheimer’s diagnoses compared to their non-HIV counterparts.
- RT inhibitors, initially developed for HIV, might inhibit similar enzymes in the brain, suggesting a potential mechanism for their effect on Alzheimer’s disease.
- The research was supported by notable foundations and the NIH, highlighting its credibility and the significant interest in translating these findings into new treatments for AD.
Alzheimer’s disease (AD) currently afflicts nearly seven million people in the U.S. With this number expected to grow to nearly 13 million by 2050, the lack of meaningful therapies represents a major unmet medical need. Scientists at Sanford Burnham Prebys have now identified promising real-world links between common HIV drugs and a reduced incidence of AD. The study, led by Jerold Chun, M.D., Ph.D., was published in Pharmaceuticals. Chun’s new research builds on his lab’s landmark publication in Nature in 2018 that described how somatic gene recombination in neurons can produce thousands of new gene variants within Alzheimer’s disease brains. Importantly, it also revealed for the first time how the Alzheimer’s-linked gene, APP, is recombined by using the same type of enzyme found in HIV. The enzyme, called reverse transcriptase (RT), copies RNA molecules and changes them into complementary DNA duplicates that can then be inserted back into DNA, producing permanent sequence changes within the cell’s DNA blueprint. HIV and many other viruses rely on RT to hijack a host’s cells to establish a chronic infection, so drugs that block the RT enzyme’s activity have become a common part of treatment cocktails for keeping HIV at bay. The brain appears to have its own RTs that are different from those in viruses, and the research team wondered if inhibiting brain RTs with HIV drugs actually helps AD patients. To assess the link between real-world RT inhibitor exposure and AD in humans, the team analyzed anonymized medical records with prescription claims from more than 225,000 control and HIV-positive patients, and found that RT inhibitor exposure was associated with a statistically significant reduced incidence and prevalence of AD. “Thus, we looked at HIV-positive individuals taking RT inhibitors and other combined antiretroviral therapies as they aged, and asked the question: How many of them got Alzheimer’s disease?” says Chun. “And the answer is that there were many fewer than might have been expected compared to the general population.” Of the more than 225,000 individuals with claims data in the study, just shy of 80,000 were HIV-positive individuals over the age of 60. More than 46,000 had taken RT inhibitors during a nearly three-year observation period from 2016 to 2019. The data was obtained through a collaboration with health information technology and clinical research firm IQVIA, led by Tiffany Chow, M.D. In living persons with HIV, there were 2.46 Alzheimer’s disease diagnoses per 1,000 persons among HIV-positive individuals taking these inhibitors, versus 6.15 for the general population. This control group was represented by more than 150,000 HIV-negative patients over the age of 60 with medical insurance claims related to treatment for the common cold. “You cannot feasibly run a prospective clinical trial with this number of patients,” Chun adds. “This approach is a way to look at how a drug can act on a large patient population.” Chun underscores that the drugs patients took in this retrospective study were designed to counter RT activity in HIV and likely only had a limited effect on many different possible forms of the enzyme active in the brain. “What we’re looking at now is very crude,” says Chun. “The clear next step for our lab is to identify which versions of RTs are at work in the AD brain so that more targeted treatments can be discovered, while prospective clinical trials of currently available RT inhibitors on persons with early AD should be pursued.” Jerold Chun, M.D. Ph.D., is a professor in the Center for Genetic Disorders and Aging Research at Sanford Burnham Prebys. Additional authors on the study include Tiffany W. Chow, Mark Raupp, Matthew W. Reynolds, Siying Li and Gwendolyn E. Kaeser. Funding: The work was supported by the National Institute on Aging – NIH (R01AG071465, R01AG065541 and R56AG073965), the Shaffer Family Foundation and the Bruce Ford & Anne Smith Bundy Foundation. Research published in Pharmaceuticals (March 22, 2024): https://doi.org/10.3390/ph17040408

|
Scooped by
Juan Lama
|
The U.S. must act decisively, acknowledging the potential gravity of H5N1 mammalian transmission. Humans have no natural immunity to the bird flu virus. The recent detection of H5N1 bird flu in U.S. cattle, coupled with reports of a dairy worker contracting the virus, demands a departure from the usual reassurances offered by federal health officials. While they emphasize there’s no cause for alarm and assert diligent monitoring, it’s imperative we break from this familiar script. H5N1, a strain of the flu virus known to infect bird species globally and several mammalian species in the U.S. since 2022, has now appeared to have breached a new barrier of inter-mammalian transmission, as exemplified by the expanding outbreak in dairy cows in several jurisdictions linked to an initial outbreak in Texas. Over time, continued transmission among cattle is likely to yield mutations that will further increase the efficiency of mammal-to-mammal transmission. As the Centers for Disease Control continues to investigate, this evolutionary leap, if confirmed, underscores the adaptability of the H5N1 virus and raises concerns about the next step required for a pandemic: its potential to further evolve for efficient human transmission. Because humans have no natural immunity to H5N1, the virus can be particularly lethal to them. Despite assertions of an overall low risk of H5N1 infection to the general population, the reality is that the understanding of this risk is limited, and it’s evolving alongside the virus. The situation could change very quickly, so it is important to be prepared. Comparisons to seasonal flu management underestimate the unique challenges posed by H5N1. Unlike its seasonal counterparts, vaccines produced and stockpiled to tackle bird flu were not designed to match this particular strain and are available in such limited quantities that they could not make a dent in averting or mitigating a pandemic, even if deployed in the early stages to dairy workers. The FDA-approved H5N1 vaccines — licensed in 2013, 2017, and 2020 — do not elicit a protective immune response after just one dose. Even after two doses, it is unknown whether the elicited immune response is sufficient to protect against infection or severe disease, as these vaccines were licensed based on their ability to generate an immune response thought to be helpful in preventing the flu. Early studies done by mRNA vaccine companies on seasonal flu are promising, which could be good news here since mRNA vaccines can be made more quickly than vaccines using eggs or cells. Congressional funding is needed to catalyze rapid vaccine development and production. While FDA-approved antiviral drugs like Tamiflu and Xofluza could be an important line of defense against H5N1, logistical barriers impede their timely administration, as they work best when given as early as possible within 48 hours of the onset of symptoms. Most Americans would find it challenging to get a prescription filled for these medicines within the optimal time frame. Streamlining access to stockpiled antiviral drugs through improved test-to-treat measures like behind-the-counter distribution or dedicated telemedicine consultations could vastly improve their effectiveness as a frontline defense. Making plans to do that need to start now. For vulnerable people — older adults and anyone who is immunocompromised — clinicians have become accustomed to relying on monoclonal antibodies. Sadly, their performance for flu has been disappointing in many clinical trials and can’t be counted on. The need for robust diagnostic capabilities cannot be overstated. H5N1 will not be detected by the typical rapid flu antigen tests that are administered in emergency rooms and many doctors’ offices. New tests will have to be made from scratch. The dismantling of diagnostic infrastructure post-Covid-19 and supply chain disruptions, however, pose significant challenges to the availability of such tests. Rapid investment in diagnostic testing, coupled with efforts to secure essential materials, is imperative to ensure timely detection and antiviral treatment. President Biden’s emphasis on infrastructure presents a unique opportunity to fortify America’s defenses against infectious diseases. A national initiative to enhance indoor air quality in schools and communal spaces could mitigate transmission risks should this virus learn how to efficiently be transmitted between humans, and would pay dividends every respiratory virus season and for years to come. In the face of uncertainty, complacency is not an option. The U.S. must act decisively, acknowledging the potential gravity of the H5N1 situation while leveraging every available resource to safeguard public health. The stakes are too high to repeat past mistakes. Luciana Borio is an infectious disease physician, a senior fellow for global health at the Council on Foreign Relations, a venture partner at ARCH Venture Partners, and former director for medical and biodefense preparedness policy at the National Security Council. Phil Krause is a virologist, infectious disease physician, and former deputy director of the Office of Vaccines Research and Review at the FDA. The authors have no links to any companies producing or evaluating any of the vaccines or therapies mentioned in this article, and declare no conflicts of interest.

|
Scooped by
Juan Lama
|
Highlights - Phylodynamic models reveal swift early mpox spread between five global regions
- Extensive, underdetected dissemination promoted rapid local transmission
- Later mpox introductions played a negligible role in prolonging regional epidemics
- N. America epidemic declined before 10% of high-risk group had vaccine-induced immunity
Summary The World Health Organization declared mpox a public health emergency of international concern in July 2022. To investigate global mpox transmission and population-level changes associated with controlling spread, we built phylogeographic and phylodynamic models to analyze MPXV genomes from five global regions together with air traffic and epidemiological data. Our models reveal community transmission prior to detection, changes in case reporting throughout the epidemic, and a large degree of transmission heterogeneity. We find that viral introductions played a limited role in prolonging spread after initial dissemination, suggesting that travel bans would have had only a minor impact. We find that mpox transmission in North America began declining before more than 10% of high-risk individuals in the USA had vaccine-induced immunity. Our findings highlight the importance of broader routine specimen screening surveillance for emerging infectious diseases and of joint integration of genomic and epidemiological information for early outbreak control. Published in Cell (Feb. 29, 2024):

|
Scooped by
Juan Lama
|
Experts warn that lax regulations could also see the virus spread to US pig farms, with serious consequences for human health. Fears are growing that the H5N1 outbreak among cattle in the United States could have been caused by contaminated animal feed. In contrast to Britain and Europe, American farmers are still allowed to feed cattle and other farm animals ground-up waste from other animals including birds. Dairy cows across six US states – and at least one farm worker – have become infected with the highly pathogenic virus, which has already killed millions of animals across the globe since 2021. The farm worker, who is thought to have been exposed via infected cattle in Texas, is only the second recorded human H5N1 case in the US. Since February, the US has investigated and discounted a further 8,000 possible exposures, according to Dr Joshua Mott, WHO senior advisor on influenza. The development is of concern because it allows the virus, which has killed millions of birds and wild mammals around the world, more opportunities to mutate...

|
Scooped by
Juan Lama
|
A man in the Mekong Delta's Tien Giang Province has become the first person in Vietnam to be infected with avian flu A/H9. The 37-year-old man, who lives in Chau Thanh District, had a fever and purchased medicine to treat himself on March 10. As his conditions got serious, he was taken to the HCMC Hospital for Tropical Diseases on March 16, where doctors diagnosed him with severe viral pneumonia.A man in the Mekong Delta's Tien Giang Province tested positive with avian flu A/H9, becoming the first recorded case in Vietnam with the disease. Test results revealed that he had influenze A, and there were genetic segments similar to the A/H9 influenze strain. The Pasteur Institute of HCMC also confirmed the result. The man is under intensive care. The man lives near a wet market that sells poultry, and there is no sick or dead animal in the areas around the patient's family. Those who have had close contact with the patient have been listed and being monitored, and they have shown no signs of respiratory infection. No outbreaks have been recorded near the patient's neighborhood. The A/H9 influenza is an avian flu infectious to people. There have been no evidence to prove that the virus can be transmitted between humans. The Ministry of Health said the man from Tien Giang is the first recorded case of A/H9 in Vietnam. From 2015 to 2023, 98 such cases have been recorded within the West Pacific region, with two deaths. There is currently no cure or vaccine for avian flu. Death rates for avian flu may reach 50%, while the rates for normal flu is at 1-4%.

|
Scooped by
Juan Lama
|
Health Alert Network (HAN). Provided by the Centers for Disease Control and Prevention (CDC). Summary The Centers for Disease Control and Prevention (CDC) is issuing this Health Alert Network (HAN) Health Advisory to inform clinicians, state health departments, and the public of a recently confirmed human infection with highly pathogenic avian influenza (HPAI) A(H5N1) virus in the United States following exposure to presumably infected dairy cattle. The U.S. Department of Agriculture (USDA) recently reported detections of highly pathogenic avian influenza A(H5N1) virus in U.S. dairy cattle in multiple states. This Health Advisory also includes a summary of interim CDC recommendations for preventing, monitoring, and conducting public health investigations of potential human infections with HPAI A(H5N1) virus. Background
A farm worker on a commercial dairy farm in Texas developed conjunctivitis on approximately March 27, 2024, and subsequently tested positive for HPAI A(H5N1) virus infection. HPAI A(H5N1) viruses have been reported in the area’s dairy cattle and wild birds. There have been no previous reports of the spread of HPAI viruses from cows to humans. The patient reported conjunctivitis with no other symptoms, was not hospitalized, and is recovering. The patient was recommended to isolate and received antiviral treatment with oseltamivir. Illness has not been identified in the patient’s household members, who received oseltamivir for post-exposure prophylaxis per CDC Recommendations for Influenza Antiviral Treatment and Chemoprophylaxis. No additional cases of human infection with HPAI A(H5N1) virus associated with the current infections in dairy cattle and birds in the United States, and no human-to-human transmission of HPAI A(H5N1) virus have been identified.
CDC has sequenced the influenza virus genome identified in a specimen collected from the patient and compared it with HPAI A(H5N1) sequences from cattle, wild birds, and poultry. While minor changes were identified in the virus sequence from the patient specimen compared to the viral sequences from cattle, both cattle and human sequences lack changes that would make them better adapted to infect mammals. In addition, there were no markers known to be associated with influenza antiviral drug resistance found in the virus sequences from the patient’s specimen, and the virus is closely related to two existing HPAI A(H5N1) candidate vaccine viruses that are already available to manufacturers, and which could be used to make vaccine if needed. This patient is the second person to test positive for HPAI A(H5N1) virus in the United States. The first case was reported in April 2022 in Colorado in a person who had contact with poultry that was presumed to be infected with HPAI A(H5N1) virus. Currently, HPAI A(H5N1) viruses are circulating among wild birds in the United States, with associated outbreaks among poultry and backyard flocks and sporadic infections in mammals....

|
Scooped by
Juan Lama
|
Laboratory-acquired infections (LAIs) and accidental pathogen escape from laboratory settings (APELS) are major concerns for the community. A risk-based approach for pathogen research management within a standard biosafety management framework is recommended but is challenging due to reasons such as inconsistency in risk tolerance and perception. Here, we performed a scoping review using publicly available, peer-reviewed journal and media reports of LAIs and instances of APELS between 2000 and 2021. We identified LAIs in 309 individuals in 94 reports for 51 pathogens. Eight fatalities (2·6% of all LAIs) were caused by infection with Neisseria meningitidis (n=3, 37·5%), Yersinia pestis (n=2, 25%), Salmonella enterica serotype Typhimurium (S Typhimurium; n=1, 12·5%), or Ebola virus (n=1, 12·5%) or were due to bovine spongiform encephalopathy (n=1, 12·5%). The top five LAI pathogens were S Typhimurium (n=154, 49·8%), Salmonella enteritidis (n=21, 6·8%), vaccinia virus (n=13, 4·2%), Brucella spp (n=12, 3·9%), and Brucella melitensis (n=11, 3·6%). 16 APELS were reported, including those for Bacillus anthracis, SARS-CoV, and poliovirus (n=3 each, 18·8%); Brucella spp and foot and mouth disease virus (n=2 each, 12·5%); and variola virus, Burkholderia pseudomallei, and influenza virus H5N1 (n=1 each, 6·3%). Continual improvement in LAI and APELS management via their root cause analysis and thorough investigation of such incidents is essential to prevent future occurrences. The results are biased due to the reliance on publicly available information, which emphasises the need for formalised global LAIs and APELS reporting to better understand the frequency of and circumstances surrounding these incidents. Published in The Lancet Microbre (Feb. 2024): https://doi.org/10.1016/S2666-5247(23)00319-1
|

|
Scooped by
Juan Lama
|
Background Growing evidence suggests that symptoms associated with post-COVID-19 condition (also known as long COVID) can affect multiple organs and systems in the human body, but their association with viral persistence is not clear. The aim of this study was to investigate the persistence of SARS-CoV-2 in diverse tissues at three timepoints following recovery from mild COVID-19, as well as its association with long COVID symptoms. Methods This single-centre, cross-sectional cohort study was done at China–Japan Friendship Hospital in Beijing, China, following the omicron wave of COVID-19 in December, 2022. Individuals with mild COVID-19 confirmed by PCR or a lateral flow test scheduled to undergo gastroscopy, surgery, or chemotherapy, or scheduled for treatment in hospital for other reasons, at 1 month, 2 months, or 4 months after infection were enrolled in this study. Residual surgical samples, gastroscopy samples, and blood samples were collected approximately 1 month (18–33 days), 2 months (55–84 days), or 4 months (115–134 days) after infection. SARS-CoV-2 was detected by digital droplet PCR and further confirmed through RNA in-situ hybridisation, immunofluorescence, and immunohistochemistry. Telephone follow-up was done at 4 months post-infection to assess the association between the persistence of SARS-CoV-2 RNA and long COVID symptoms. Findings Between Jan 3 and April 28, 2023, 317 tissue samples were collected from 225 patients, including 201 residual surgical specimens, 59 gastroscopy samples, and 57 blood component samples. Viral RNA was detected in 16 (30%) of 53 solid tissue samples collected at 1 month, 38 (27%) of 141 collected at 2 months, and seven (11%) of 66 collected at 4 months. Viral RNA was distributed across ten different types of solid tissues, including liver, kidney, stomach, intestine, brain, blood vessel, lung, breast, skin, and thyroid. Additionally, subgenomic RNA was detected in 26 (43%) of 61 solid tissue samples tested for subgenomic RNA that also tested positive for viral RNA. At 2 months after infection, viral RNA was detected in the plasma of three (33%), granulocytes of one (11%), and peripheral blood mononuclear cells of two (22%) of nine patients who were immunocompromised, but in none of these blood compartments in ten patients who were immunocompetent. Among 213 patients who completed the telephone questionnaire, 72 (34%) reported at least one long COVID symptom, with fatigue (21%, 44 of 213) being the most frequent symptom. Detection of viral RNA in recovered patients was significantly associated with the development of long COVID symptoms (odds ratio 5·17, 95% CI 2·64–10·13, p<0·0001). Patients with higher virus copy numbers had a higher likelihood of developing long COVID symptoms. Interpretation Our findings suggest that residual SARS-CoV-2 can persist in patients who have recovered from mild COVID-19 and that there is a significant association between viral persistence and long COVID symptoms. Further research is needed to verify a mechanistic link and identify potential targets to improve long COVID symptoms. Published in The Lancet Infectious Diseases (April 22, 2024):

|
Scooped by
Juan Lama
|
In early March, veterinarian Barb Petersen noticed the dairy cows she cared for on a Texas farm looked sick and produced less milk, and that it was off-color and thick. Birds and cats on the farm were dying, too. Petersen contacted Kay Russo at Novonesis, a company that helps farms keep their animals healthy and productive. “I said, you know, I may sound like a crazy, tinfoil hat–wearing person,” Russo, also a veterinarian, recalled at a 5 April public talk sponsored by her company. “But this sounds a bit like influenza to me.” She was right, as Petersen and Russo soon learned. On 19 March, birds on the Texas farm tested positive for H5N1, the highly pathogenic avian flu that has been devastating poultry and wild birds around the world for more than 2 years. Two days later, tests of cow milk and cats came back positive for the same virus, designated 2.3.4.4b. “I was at Target with my kids, and a series of expletives came out of my mouth,” Russo said. Now, 3 weeks into the first ever outbreak of a bird flu virus in dairy cattle, Russo and others are still dismayed—this time by the many questions that remain about the infections and the threat they may pose to livestock and people, and by the federal response. Eight U.S. states have reported infected cows, but government scientists have released few details about how the virus is spreading. In the face of mounting criticism about sharing little genetic data—which could indicate how the virus is changing and its potential for further spread—the U.S. Department of Agriculture (USDA) did an unusual Sunday evening dump on 21 April of 202 sequences from cattle into a public database. (Some may be different samples from the same animal.) And despite public reassurances about the safety of the country’s milk supply, officials have yet to provide supporting data. The cow infections, first confirmed by USDA on 25 March, were “a bit of an inconvenient truth,” Russo says. Taking and testing samples, sequencing viruses, and running experiments can take days, if not weeks, she acknowledges. But she thinks that although officials are trying to protect the public, they are also hesitant to cause undue harm to the dairy and beef industries. “There is a fine line of respecting the market, but also allowing for the work to be done from a scientific perspective,” Russo says. Only one human case linked to cattle has been confirmed to date, and symptoms were limited to conjunctivitis, also known as pink eye. But Russo and many other vets have heard anecdotes about workers who have pink eye and other symptoms—including fever, cough, and lethargy—and do not want to be tested or seen by doctors. James Lowe, a researcher who specializes in pig influenza viruses, says policies for monitoring exposed people vary greatly between states. “I believe there are probably lots of human cases,” he says, noting that most likely are asymptomatic. Russo says she is heartened that the Centers for Disease Control and Prevention has “really started to mobilize and do the right thing,” including linking with state and local health departments, as well as vets, to monitor the health of workers on affected farms. As farms in more states began to report infected dairy cows last month, one theory held migratory birds were carrying the virus across the country and introducing it repeatedly to different dairy herds. But Lowe dismisses the idea as “some fanciful thinking by some cow veterinarians and some cow producers” who hoped to “protect the industry.” Richard Webby, an avian influenza researcher at St. Jude Children’s Research Hospital, notes that available data on the virus’ genetic sequence show “no smoking guns”—mutations that could enable it to jump readily from birds to cows. At a 4 April meeting organized by a group known as the Global Framework for the Progressive Control of Transboundary Animal Diseases, Suelee Robbe Austerman of USDA’s National Veterinary Services Laboratory said “a single spillover event or a couple of very closely related spillover events” from birds is more likely. The cow virus—which USDA has designated 3.13—could then have moved between farms as southern herds were moved northward for the spring, perhaps spreading from animal to animal on milking equipment. Lowe notes that researchers at Iowa State necropsied infected cows and found no virus in their respiratory tracts, which could enable spread through the air. USDA considers H5N1 in poultry a “program disease,” which means it is foreign to the country and subject to regulations to control it. But the agency has made no such determination for the cow disease, which means the federal government has no power to restrict movement of cattle or to require testing and reporting of infected herds or of the humans who work with them. As a result, confusion is rife. A knowledgeable source who asked not to be identified says cattle that were healthy when they left a Texas farm appear to have brought the virus to a North Carolina farm. That raises the possibility that many cattle are infected but asymptomatic, which would make the virus harder to contain. Evidence suggests it has spread from cattle back to poultry, USDA says. Another source told Sciencenone that birds tested positive for the 3.13 strain and were culled at a Minnesota turkey farm right next to a dairy farm that refused to test its cattle. The industry and veterinarians bear some responsibility for the confusion, Lowe says. “We didn’t even try to get ahead of this thing,” he says. “That’s a black mark on the industry and on the profession.” Webby also faults USDA for not releasing data more quickly. The agency made six sequences from cattle—plus six related ones from birds and one from a skunk—available on the GISAID database on 29 March, 1 week after learning that cows were infected. It released one more sequence on 5 April, but then shared nothing else until the data dump 16 days later. Webby suspects the agency has moved cautiously because of the potential impact on the dairy industry. “There are a lot of people who are vested in this, and their livelihoods, at least on paper, can be impacted by whatever is found.” Thijs Kuiken, an avian influenza researcher at Erasmus Medical Center, says the “very sparse” information released by the U.S. government has international implications, too. State and federal animal health authorities have “abundant information … that [has] not been made public, but would be informative for health professionals and scientists” in the United States and abroad, he says, “to be able to better assess the outbreak and take measures, both for animal health and for human health.” He notes that even the new sequences released by USDA do not include locations of the samples or the date they were taken. USDA said in a statement to Sciencenone that it will continue “to provide timely and accurate updates” and that its staff “are working around the clock” on a thorough epidemiological investigation to determine routes of transmission and help control spread. With its Sunday evening release, USDA made the unusual move to bypass GISAID—which requires a login and restricts how data can be used—and instead posted on a database run by the National Institutes of Health that everyone can access and do with as they please. USDA explained it did this “in the interest of public transparency and ensuring the scientific community has access to this information as quickly as possible to encourage disease research.” The dairy industry is particularly worried that sales could suffer because of unwarranted fears that milk might infect people with the virus. The Food and Drug Administration (FDA) has said it has “no concern” that pasteurized milk presents risks because the process “has continually proven to inactivate bacteria and viruses, like influenza, in milk.” But Lowe says for this specific virus, there are no data to back that claim. That’s “shocking,” he says. “It should not be that hard to get that data. I do not know why the foot dragging is occurring there.” In a statement to Sciencenone, FDA said the agency is working to “reinforce our current assessment that the commercial milk supply is safe” and will share data as soon as possible. “U.S. government partners are working with all deliberate speed on a wide range of studies looking at milk along all stages of production, including on the farm, during processing and on shelves,” the statement said. Daniel Perez, an influenza researcher at the University of Georgia, is doing his own test tube study of pasteurization of milk spiked with a different avian influenza virus. The fragile lipid envelope surrounding influenza viruses should make them vulnerable, he says. Still, he wonders whether the commonly used “high temperature, short time” pasteurization, which heats milk to about 72°C for 15 or 20 seconds, is enough to inactivate all the virus in a sample. Lowe predicts that with increased safety precautions at farms, including more rigorous cleaning of milking equipment, and the end to the seasonal movement of cattle, this outbreak likely will die out—as often happens when bird influenza viruses infect mammals that do not make particularly good hosts. But he and other researchers say to truly assess the continuing threat of H5N1 in cattle, they need to understand how this virus succeeded at infecting dairy cows in the first place. “Which cells is the virus replicating in? How is it getting around the body? What receptor is it using to get into cells?” Lowe wonders. “Those are the really interesting questions.” Correction, 23 April 2024, 7:35 a.m.: A previous version of this story said James Lowe necropsied infected cows. He didn't but noted that researchers at Iowa State University did.

|
Scooped by
Juan Lama
|
Significance The recent Covid-19 pandemic highlighted that understanding the channels of disease transmission is crucially important for public health policies. However, measuring transmissions occurring through casual contact in the public space is highly challenging as researchers generally do not observe when infected individuals intersect casually with noninfected individuals. We overcome this methodological challenge in the context of the Covid-19 pandemic by combining card payment data, indicating exactly where and when individuals visited stores, with test data indicating when they were infected. We document that exposure to an infected individual in a store is associated with a significantly higher infection rate in the following week. Our estimates imply that transmissions between retail shoppers made a substantial contribution to the Covid-19 pandemic. Abstract This paper presents quasiexperimental evidence of Covid-19 transmission through casual contact between customers in retail stores. For a large sample of individuals in Denmark, we match card payment data, indicating exactly where and when each individual made purchases, with Covid-19 test data, indicating when each individual was tested and whether the test was positive. The resulting dataset identifies more than 100,000 instances where an infected individual made a purchase in a store and, in each instance, allows us to track the infection dynamics of other individuals who made purchases in the same store around the same time. We estimate transmissions by comparing the infection rate of exposed customers, who made a purchase within 5 min of an infected individual, and nonexposed customers, who made a purchase in the same store 16 to 30 min before. We find that exposure to an infected individual in a store increases the infection rate by around 0.12 percentage points (P < 0.001) between day 3 and day 7 after exposure. The estimates imply that transmissions in stores contributed around 0.04 to the reproduction number for the average infected individual and significantly more in the period where Omicron was the dominant variant. Published in PNAS (April 17, 2024):

|
Scooped by
Juan Lama
|
New research presented at the ESCMID Global Congress (formerly ECCMID) in Barcelona, Spain (27–30 April) shows that in a VACCELERATE Consortium survey study in which infectious diseases experts were asked to rank pathogens in order of their pandemic potential, influenza was considered the pathogen of highest pandemic risk, with 57% ranking influenza as number one, and a further 17% ranking it second. The study is by Dr. Jon Salmanton-García, University of Cologne, Faculty of Medicine and University Hospital Cologne, Institute of Translational Research, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), Cologne, Germany, and colleagues from across Europe, and published in the journal Travel Medicine and Infectious Disease. Other highly ranked pathogens included Disease X (as yet unknown disease) with 21% ranking this number one and 14% second. SARS-CoV-2 was third in terms of number one responses on 8%, with 16% voting it number two, while the original SARS-CoV virus that circulated in 2002–03 was voted number one by 2% of respondents and second by 8%. Crimean-Congo hemorrhagic fever virus (CCHF virus) and Ebola virus were joint fifth, with 1.6% respondents voting them first. Nipah virus, henipavirus, and Rift Valley fever virus were among the pathogens ranked lowest in terms of their perceived pandemic potential. The World Health Organization (WHO) has outlined a comprehensive Research and Development (R&D) Blueprint for Action to Prevent Epidemics, focusing on key infectious diseases that pose significant threats to public health. These diseases were selected after rigorous evaluation, taking into account factors like transmissibility, infectivity, severity, and their potential for evolution. In alignment with the WHO's R&D Blueprint, the VACCELERATE Site Network (a pan-European network of sites collaborating on COVID-19, other infectious diseases and general pandemic preparedness infectious diseases) engaged infectious disease experts from around the world, both among and outside its members, to rank the diseases listed in terms of their perceived risk of instigating a pandemic. Participants were tasked with ranking various pathogens based on their perceived pandemic risk, encompassing diseases featured in the WHO R&D Blueprint and additional pathogens. Experts could rank up to 14 pathogens in the order of their perceived risk (the 13 pathogens listed plus Disease X—as yet unknown pathogen) in any order, and also suggest pathogens not listed to include in their maximum of 14. Each pathogen received a score based on its positions. A total of 187 responses were collected from infectious disease experts hailing from 57 different countries Among the countries providing the highest number of responses, Germany accounted for 27 replies (14%), followed by Spain with 20 replies (11%), and Italy with 14 replies (8%). Influenza viruses emerged as the most concerning pathogen with other highly ranked pathogens including Disease X, SARS-CoV-2, SARS-CoV, and the Ebola virus. Conversely, Hantavirus, Lassa virus, Nipah virus, henipavirus and Rift Valley fever virus were among the pathogens ranked lowest in terms of their pandemic potential. The authors conclude, "The study revealed that influenza, disease X, SARS-CoV-1, SARS-CoV-2, and the Ebola virus are the most worrisome pathogens concerning their pandemic potential. These pathogens are characterized by their transmissibility through respiratory droplets and a history of previous epidemic or pandemic outbreaks." Commenting on the top ranking of influenza, Dr. Salmanton-García adds, "Each winter we have an influenza season. One could say that this means that every winter there are little pandemics. They are more or less controlled because the different strains are not virulent enough. Yet, every season the strains involved change, that is the reason why we can get influenza several times in life and vaccines change year to year. In case a new strain becomes more virulent, this control could be lost." But he adds the world is now much more prepared due to the COVID-19 pandemic, whereas before a lot of the focus had been on a potential influenza pandemic. He says, "In COVID-19 pandemic we have learned many things on how to approach a respiratory virus pandemic. This includes social distancing, hand cleaning, face masks, a renewed focus on vaccination, and trust in health care institutions. In parallel, institutions have also learnt a lot. Preparedness and surveillance are now, vitally, better-funded." Study published in Travel Medicine and Infectious Disease (Feb. 2024): https://doi.org/10.1016/j.tmaid.2023.102676

|
Scooped by
Juan Lama
|
The 20-month-long infection is the longest known and highlights how prolonged infections enable the pandemic virus to accumulate genetic changes—potentially spawning new variants of concern. A Covid-19 patient with a weakened immune system incubated a highly mutated novel strain over 613 days before succumbing to an underlying illness, researchers in the Netherlands found. The patient, a 72-year-old man with a blood disorder, failed to mount a strong immune response to multiple Covid shots before catching the omicron variant in February 2022. Detailed analysis of specimens collected from more than two dozen nose and throat swabs found the coronavirus developed resistance to sotrovimab, a Covid antibody treatment, within a few weeks, scientists at the University of Amsterdam’s Centre for Experimental and Molecular Medicine said. It later acquired over 50 mutations, including some that suggested an enhanced ability to evade immune defenses, they said. The 20-month-long SARS-CoV-2 infection is the longest known, according to the researchers, who are presenting the case at a medical meeting in Barcelona next week. While his mutant virus wasn’t known to have infected other people, it highlights how prolonged infections enable the pandemic virus to accumulate genetic changes, potentially spawning new variants of concern. “This case underscores the risk of persistent SARS-CoV-2 infections in immunocompromised individuals,” the authors said. “We emphasize the importance of continuing genomic surveillance of SARS-CoV-2 evolution in immunocompromised individuals with persistent infections.” Scientists studying genomic data collected from wastewater samples have reported evidence of individuals in the community shedding heavily mutated coronaviruses for more than four years. Such persistent infections may also be causing patients to experience long Covid symptoms, research suggests.

|
Scooped by
Juan Lama
|
New research has uncovered a social world of viruses full of cheating, cooperation and other intrigues, suggesting that viruses make sense only as members of a community. Ever since viruses came to light in the late 1800s, scientists have set them apart from the rest of life. Viruses were far smaller than cells, and inside their protein shells they carried little more than genes. They could not grow, copy their own genes or do much of anything. Researchers assumed that each virus was a solitary particle drifting alone through the world, able to replicate only if it happened to bump into the right cell that could take it in. This simplicity was what attracted many scientists to viruses in the first place, said Marco Vignuzzi, a virologist at the Singapore Agency for Science, Research and Technology Infectious Diseases Labs. “We were trying to be reductionist.” That reductionism paid off. Studies on viruses were crucial to the birth of modern biology. Lacking the complexity of cells, they revealed fundamental rules about how genes work. But viral reductionism came at a cost, Vignuzzi said: By assuming viruses are simple, you blind yourself to the possibility that they might be complicated in ways you don’t know about yet....

|
Scooped by
Juan Lama
|
A new lineage of the mpox virus linked to efficient human-to-human transmission has been identified in the Democratic Republic of the Congo (DRC) — and the researchers behind the finding are calling for swift action to 'avert another global mpox outbreak.' A team of Canadian and international scientists working on the ground in DRC began tracking a large mpox outbreak in the country's Kamituga mining region last year. From October 2023 onward, cases spread rapidly with more than 240 suspected infections identified within five months. Out of roughly 100 confirmed cases, a third were among sex workers, the team wrote in their latest paper. Mpox — previously known as monkeypox — burst onto the global landscape in 2022, spreading to dozens of countries through sexual networks that largely impacted men who have sex with men. Infections can lead to painful lesions and, in more severe cases, sepsis, lung nodules and even death. Genomic analysis of the recent Kamituga outbreak uncovered what lead researcher Dr. Placide Mbala-Kingebeni calls an "alarming" finding: a new, distinct clade Ib lineage of the mpox virus featuring mutations that are a hallmark of efficient transmission between humans. Mbala-Kingebeni's findings were published online this week as a pre-print, meaning the research has not yet been peer-reviewed. The researchers say the ongoing situation in Kamituga echoes the 2017 to 2018 outbreak of clade IIb mpox in Nigeria, which is now widely considered a harbinger to the unprecedented global spread of the disease a few years later. Mbala-Kingbeni told CBC News the worldwide spread of mpox in 2022 involved a form of the virus that's thought to cause less severe disease, though some scientists warn underreporting of cases makes it tricky to know the precise differences between each clade. (Clades are subtypes of a virus that arise from a common ancestor. In the case of mpox, the two main clades identified so far are clade I and clade II.) "There is a high risk that this more virulent clade, that's more adapted to human-to-human transmission ... can be transmitted silently from one person to another and be sustained at a global level," added Mbala-Kingbeni, an associate professor at the University of Kinshasa and head of epidemiology and global health at the DRC National Institute of Biomedical Research's Clinical Research Center. Urgent measures are needed — including expanded surveillance, contact tracing and targeted vaccination efforts — to contain this "pandemic-potential" outbreak, the researchers wrote. Ongoing DRC outbreaks 'alarming' The team's concern stems, in part, from Kamituga's demographics. The densely-populated region lacks healthcare infrastructure to handle a large-scale outbreak, and the researchers believe the reported cases are likely an underestimate. It's also a travel hub, with large amounts of movement between the region and neighbouring countries in Africa, such as Rwanda and Burundi, including sex workers who "frequently return to their countries of origin." "The highly mobile nature of this mining population poses a substantial risk of outbreak escalation beyond the current area and across borders," reads the paper. Toronto-based infectious diseases specialist Dr. Isaac Bogoch, who was not involved in the research, praised the team for showing a "subtle but real" genetic difference in the mpox virus samples from Kamituga. The research further raises alarms about the record-breaking levels of mpox circulating in DRC, he added, and the country's lack of vaccines or therapeutics to contain the spread of the virus. But he cautioned against over-interpreting the results, saying the findings don't prove this new lineage is even more transmissible than before or capable of evading existing treatments and vaccines. Further research is needed, Bogoch stressed, to better understand a virus that's causing massive outbreaks in Africa and is still quietly circulating locally in Canada as well — with more than 20 cases reported in Toronto alone so far this year. The study also doesn't change the existing possibility that a different lineage of mpox could eventually jump internationally, Bogoch added. "Without this mutation we would have this exact same conversation," he said. "The alarming part to me is we know there's an ongoing outbreak in [DRC], and we still don't have a coordinated global response." Preprint of the study available in medRxiv (April 15, 2024); https://doi.org/10.1101/2024.04.12.24305195

|
Scooped by
Juan Lama
|
Abstract
SARS-CoV-2 has been shown to cause wide-ranging ocular abnormalities and vision impairment in COVID-19 patients. However, there is limited understanding of SARS-CoV-2 in ocular transmission, tropism, and associated pathologies. The presence of viral RNA in corneal/conjunctival tissue and tears, along with the evidence of viral entry receptors on the ocular surface, has led to speculation that the eye may serve as a potential route of SARS-CoV-2 transmission. Here, we investigated the interaction of SARS-CoV-2 with cells lining the blood-retinal barrier (BRB) and the role of the eye in its transmission and tropism. The results from our study suggest that SARS-CoV-2 ocular exposure does not cause lung infection and moribund illness in K18-hACE2 mice despite the extended presence of viral remnants in various ocular tissues. In contrast, intranasal exposure not only resulted in SARS-CoV-2 spike (S) protein in different ocular tissues but also induces a hyperinflammatory immune response in the retina.
Additionally, the long-term exposure to viral S-protein caused microaneurysm, retinal pigmented epithelium (RPE) mottling, retinal atrophy, and vein occlusion in mouse eyes. Notably, cells lining the BRB, the outer barrier, RPE, and the inner barrier, retinal vascular endothelium, were highly permissive to SARS-CoV-2 replication. Unexpectedly, primary human corneal epithelial cells were comparatively resistant to SARS-CoV-2 infection. The cells lining the BRB showed induced expression of viral entry receptors and increased susceptibility towards SARS-CoV-2-induced cell death. Furthermore, hyperglycemic conditions enhanced the viral entry receptor expression, infectivity, and susceptibility of SARS-CoV-2-induced cell death in the BRB cells, confirming the reported heightened pathological manifestations in comorbid populations. Collectively, our study provides the first evidence of SARS-CoV-2 ocular tropism via cells lining the BRB and that the virus can infect the retina via systemic permeation and induce retinal inflammation. Author summary SARS-CoV-2 is known to cause several ocular manifestations in COVID-19 patients; however, the role of eyes in viral transmission and ocular tissue tropism remains elusive. The presence of viral remnants in various ocular tissues and fluids from COVID-19 patients has led to an assumption that SARS-CoV-2 may be transmitted through the eyes. Here, we show that SARS-CoV-2 ocular tropism is through cells lining the BRB. SARS-CoV-2 not only infects the various parts of the eye via systemic exposure but also induces a hyperinflammatory immune and antiviral response in the retina. Unexpectedly, the corneal epithelium was found to be resistant to SARS-CoV-2 infection, and ocular exposure of SARS-CoV-2 failed to cause lung pathology and moribund illness. Cells lining the BRB showed induced expression of viral entry receptors and enhanced susceptibility towards SARS-CoV-2-induced cell death, which is further potentiated with comorbidities such as hyperglycemia. Our findings from this study shed light on the role of BRB in SARS-CoV-2 ocular tropism and the role of eyes in viral transmission. Published in PLOS Pathogen (April 10, 2024): https://doi.org/10.1371/journal.ppat.1012156

|
Scooped by
Juan Lama
|
We reviewed information about mammals naturally infected by highly pathogenic avian influenza A virus subtype H5N1 during 2 periods: the current panzootic (2020–2023) and previous waves of infection (2003–2019). In the current panzootic, 26 countries have reported >48 mammal species infected by H5N1 virus; in some cases, the virus has affected thousands of individual animals. The geographic area and the number of species affected by the current event are considerably larger than in previous waves of infection. The most plausible source of mammal infection in both periods appears to be close contact with infected birds, including their ingestion. Some studies, especially in the current panzootic, suggest that mammal-to-mammal transmission might be responsible for some infections; some mutations found could help this avian pathogen replicate in mammals. H5N1 virus may be changing and adapting to infect mammals. Continuous surveillance is essential to mitigate the risk for a global pandemic. Since last century, highly pathogenic avian influenza (HPAI) viruses have caused diverse waves of infection. However, the ongoing panzootic event (2020–2023) caused by HPAI A(H5N1) virus could become one of the most important in terms of economic losses, geographic areas affected, and numbers of species and individual animals infected. This pathogen appears to be emerging in several regions of the world (e.g., South America); it has caused death in domestic and wild birds but also in mammals. This trend is of great concern because it may indicate a change in the dynamics of this pathogen (i.e., an increase in their range of hosts and the severity of the disease). H5N1 has affected several mammal species since 2003, thus raising concern because H5N1 mammalian adaptation could represent a risk not only for diverse wild mammals but also for human health. Unfortunately, information about this topic, especially related to the current panzootic (2020–2023), is disperse and available often only in gray literature (e.g., databases and official government websites). This fact complicates access and evaluation for many stakeholders working on the front lines (e.g., wildlife managers, conservationists, and public health authorities at regional and local levels). For this article, we compiled and analyzed information from scientific literature about mammal species, including humans, naturally affected by the current panzootic event and compared those findings with the outcomes of previous waves of H5N1 infection. We focus particularly on the species infected, their habitat, phylogeny, and trophic level, and the sources of infection, virus mutations, clinical signs, and necropsy findings associated with this virus. We also address potential risks for biodiversity and human health. Published in Emerging Infectious Diseases (March 2024): https://doi.org/10.3201/eid3003.231098

|
Scooped by
Juan Lama
|
Lancet study solidifies evidence of long-term viral persistence after COVID; demonstrates urgency of sustained research into the chronic health consequence of the virus. Medford MA, April 8, 2024 – Research published today in Lancet Infectious Disease and supported by PolyBio Research Foundation provides the strongest evidence yet that the COVID virus can persist for months or years after infection. The findings, published by a UC San Francisco/Harvard Medical School team, found that proteins created by the virus were still present for up to 14 months in a quarter of people tested. This demonstrates SARS-CoV-2 viral persistence as an urgent area of research underlying a breadth of chronic disease after COVID. “The fact that every new SARS-CoV-2 infection has the potential to become chronic is perhaps the single most concerning aspect of this virus” says Dr. Amy Proal, President of PolyBio. “We have compelling data that viral persistence is much more common than recognized which could have major health implications.” Solid Evidence of Persistent Infection The researchers analyzed blood samples from 171 people who had been infected with the virus. Using a novel ultra-sensitive blood test, they found that proteins from the virus were still present up to one year after infection in up to 25% of people. These findings greatly bolster evidence that the coronavirus can linger in tissue and organs, even after recovery from acute infection. The likelihood of detecting the COVID proteins indicative of persistent virus was about twice as high among those who were hospitalized for COVID as it was for those who were not. It was also higher for those who were not hospitalized but reported being sicker. The researchers note that viral proteins identified in the study could not have been a result the COVID vaccine, since nearly all study participants had not received the shots prior to the blood collection. In addition, proteins were only rarely found in banked blood obtained from over 200 people before the pandemic, confirming the accuracy of the blood test used in the research. “Finding COVID-19 proteins floating in the bloodstream for more than a year following initial infection was a surprise to most of us and is the product of new molecular tools. This finding now firmly informs the next step in our research agenda — is persistence of the SARS-CoV-2 virus responsible for causing people’s current symptoms in Long COVID or medical events they may have in the future?” says Jeffrey N. Martin, Head of the Division of Clinical Epidemiology at University of California, San Francisco who co-led the study. Martin is part of a multi-disciplinary collaboration behind the findings. The blood test used to find viral proteins was created by Harvard Medical School’s David Walt – founder of biotech companies that have transformed technologies across biomedical research. The blood samples were provided by volunteers in the UCSF Long-term Impact of Infection with Novel Coronavirus (LIINC) study. The UCSF team contains experts who helped make HIV/AIDS a treatable disease and pivoted their infrastructure into Long COVID via LIINC in April of 2020. The UCSF team is part of PolyBio’s LongCovid Research Consortium – a privately funded global collaborationof scientists working rapidly to document SARS-CoV-2 persistence in Long COVID, with data channeled into clinical trials. In a seminal Nature paper, the Consortium delineated a roadmap for the study of SARS-CoV-2 persistence that is being executed by dozens of international teams. However, with an estimated 18 million adults and 5.8 million children suffering from Long COVID, government investment is also needed. SARS-CoV-2 has even been found in the lymph nodes of children months after COVID, suggesting persistent infection can begin early in life. “COVID persistence could contribute to Long COVID in both adults and children” says Michael Peluso, MD, principal investigator of LIINC and an infectious disease researcher in the UCSF School of Medicine. “We must rapidly keep studying that possibility.” About PolyBio PolyBio Research Foundation is a 501(c)3 transforming how complex chronic conditions like Long COVID are studied, diagnosed, and treated. PolyBio conceptualizes research projects that identify root cause drivers of chronic conditions and builds collaborative teams to make the projects a reality. PolyBio is supported by numerous donors including Kanro – a philanthropic fund to support open source scientific research established by Vitalik Buterin, creator of Ethereum. Research published in The Lancet Infectious Diseases (April 8, 2024): https://doi.org/10.1016/S1473-3099(24)00211-1

|
Scooped by
Juan Lama
|
Hong Kong issued a major health alert as a man battles for his life in intensive care after contracting a deadly form of herpes from monkeys. A concerning health alert has been issued in Hong Kong as a 37-year-old man fights for his life in intensive care after contracting a deadly form of herpes from monkeys. The man's condition has prompted health officials to caution against interactions with wild macaques, urging the public to exercise caution. The victim sustained injuries from wild macaques in February during a visit to Kam Shan Country Park, colloquially known as 'Monkey Hill'. Initially admitted to the hospital with a fever on March 21, his condition deteriorated rapidly, leading to a transfer to the intensive care unit (ICU) where he remains in critical condition. Health authorities confirmed the man's ailment as herpes B virus, also known as herpes simiae, a disease commonly found in macaques. While this virus typically manifests as mild symptoms or remains asymptomatic in monkeys, it poses severe risks to humans, potentially leading to life-threatening complications. Symptoms of the virus in humans can escalate from fever to respiratory issues, nausea, neurological impairments such as numbness or coordination problems, and in severe cases, irreversible brain damage or fatality. Despite being first discovered in 1932, human cases remain rare, with approximately 50 recorded instances and 21 fatalities globally. Dr. Wilson Lam, president of the Hong Kong Society of Infectious Diseases, highlighted the seriousness of the situation saying: "We don’t know the virus very well, but based on the limited data, if humans are in contact with the virus, there’s a high chance of infection. It can have serious health consequences that affect the spinal nerves and central nervous system." The incident marks the first recorded case of human infection with the B virus in Hong Kong, despite the prevalence of macaques in the region. Health officials urged residents and visitors to refrain from touching or feeding wild monkeys to prevent potential transmission of the virus. As investigations continue, authorities stressed the importance of public cooperation in preventing the spread of this potentially lethal virus.

|
Scooped by
Juan Lama
|
The bird flu virus spreading through dairy cattle in the United States may be expanding its reach via milking equipment, the people doing the milking, or both, U.S. Department of Agriculture (USDA) representatives reported today at an international, virtual meeting held to update the situation. The avian virus may not be spreading directly from cows breathing on cows, as some researchers have speculated, according to USDA scientists who took part in the meeting, organized jointly by the World Organisation for Animal Health and the United Nations’s Food and Agricultural Organization. “We haven’t seen any true indication that the cows are actively shedding virus and exposing it directly to other animals,” said USDA’s Mark Lyons, who directs ruminant health for the agency and presented some of its data. The finding might also point to ways to protect humans. So far one worker at a dairy farm with infected cattle was found to have the virus, but no other human cases have been confirmed. USDA researchers tested milk, nasal swabs, and blood from cows at affected dairies and only found clear signals of the virus in the milk. “Right now, we don’t have evidence that the virus is actively replicating within the body of the cow other than the udder,” Suelee Robbe Austerman of USDA’s National Veterinary Services Laboratory told the gathering. The virus might be transmitted from cow to cow in milk droplets on dairy workers’ clothing or gloves, or in the suction cups attached to the udders for milking, Lyons said. (In a 30 March interview with Sciencenone, Thijs Kuiken, a leading avian influenza researcher at Erasmus Medical Center, had suggested the milking machines might be responsible because the components may not always be disinfected between cows.) The influenza virus causing the outbreak, an H5N1 subtype that is known as clade 2.3.4.4b, has devastated wild birds and poultry around the world for more than 2 years, and researchers at first thought migratory birds were responsible for spreading it to all of the affected dairy farms. But USDA scientists now think the movement of cattle, which are frequently transported from the southern parts of the country to the Midwest and north in the spring, may also have played an important role. And they floated the possibility, without naming specific herds or locations, that all affected cows may trace back to a single farm. Since first revealing the infection of dairy cows with this bird flu virus on 25 March, USDA has confirmed it has spread to cattle in six states. And the agency has used the virus’ genetics to track its movements. The virus found on U.S. farms has a specific genetic signature, which led USDA to name it 3.13. “It’s not a common [strain], but it’s very much a descendant of the viruses that have been dominating the flyways in the Pacific and the central flyways in the United States,” Robbe Austerman said. The USDA scientists reported that the agency’s analysis of the different viruses from the cows indicates they likely came from one source. Cows from infected farms in Texas appear to have moved the virus to farms in Idaho, Michigan, and Ohio. “The cow viruses so far have all been similar enough that it would be consistent with a single spillover event or a couple of very closely related spillover events,” Robbe Austerman said. “So far we don’t have any evidence that this is being introduced multiple times into the cows.” The virus in cows could spread to poultry, which has that industry on edge. USDA tightly regulates avian influenza virus strains that are deadly to poultry, requiring culling of entire flocks if one bird tests positive for this H5N1 or one of its relatives; to date, commercial and backyard poultry farms have had to cull 85 million birds because of this virus. But USDA is not calling for any such drastic measures with cow herds. In response to questions from Sciencenone, Ashley Peterson, who handles regulatory affairs for the National Chicken Council, said “out of an abundance of caution, we believe it is prudent to restrict the movement of cows from positive herds.” Although some researchers agree USDA should stop the transport of dairy cows, the agency has so far declined to take that disruptive action. “We heavily rely on the producers who have been isolating the animals within the dairy herds,” Lyons said. USDA also says it has no evidence of beef cattle becoming infected. USDA and the Food and Drug Administration have stressed that pasteurization kills viruses, so there is “no concern” about the safety of commercial milk. They do recommend people not consume raw milk or products made from it. Sources tell Science none some dairy farms that were later shown to be infected first noticed dead cats as early as mid-February. The animals, which often drink spilled milk on farms, “were the canary in the mine,” one said. The cows’ milk was also unusually thick. Those signs, coupled with the discovery of dead birds on the farms, led to the testing of cow milk for the bird flu virus and the 25 March USDA announcement. Oddly, the dead birds on infected farms were not waterfowl, the migratory birds that typically spread the avian flu viruses to poultry, but “peridomestic” species such as grackles, blackbirds, and pigeons. One farm had the virus detected in poultry before it was found in its cows, and although Robbe Austerman said she hesitated to call it the “ancestral strain,” she noted that “all of the detection so far in cattle are clustered around that.” Further studies should clarify how the virus wound up in cows, which sometimes become infected with flu viruses but have never before been shown to have one that causes high mortality in birds. One possibility is that birds infected cows by shedding their droppings in the cows’ feed or water. But bird flu viruses in the past have also spread for many kilometers in the wind, moving from one poultry farm to another. So the current H5N1 strain could have moved from waterfowl to poultry to cattle—or directly from poultry to cattle and then even to the peridomestic birds. “This could be a multifactorial presentation that we’re seeing,” Lyons said. “Lots of questions still to be answered.”

|
Scooped by
Juan Lama
|
BACKGROUND Nirmatrelvir in combination with ritonavir is an antiviral treatment for mild-to-moderate coronavirus disease 2019 (Covid-19). The efficacy of this treatment in patients who are at standard risk for severe Covid-19 or who are fully vaccinated and have at least one risk factor for severe Covid-19 has not been established. METHODS In this phase 2–3 trial, we randomly assigned adults who had confirmed Covid-19 with symptom onset within the past 5 days in a 1:1 ratio to receive nirmatrelvir–ritonavir or placebo every 12 hours for 5 days. Patients who were fully vaccinated against Covid-19 and who had at least one risk factor for severe disease, as well as patients without such risk factors who had never been vaccinated against Covid-19 or had not been vaccinated within the previous year, were eligible for participation. Participants logged the presence and severity of prespecified Covid-19 signs and symptoms daily from day 1 through day 28. The primary end point was the time to sustained alleviation of all targeted Covid-19 signs and symptoms. Covid-19–related hospitalization and death from any cause were also assessed through day 28. RESULTS Among the 1296 participants who underwent randomization and were included in the full analysis population, 1288 received at least one dose of nirmatrelvir–ritonavir (654 participants) or placebo (634 participants) and had at least one postbaseline visit. The median time to sustained alleviation of all targeted signs and symptoms of Covid-19 was 12 days in the nirmatrelvir–ritonavir group and 13 days in the placebo group (P=0.60). Five participants (0.8%) in the nirmatrelvir–ritonavir group and 10 (1.6%) in the placebo group were hospitalized for Covid-19 or died from any cause (difference, −0.8 percentage points; 95% confidence interval, −2.0 to 0.4). The percentages of participants with adverse events were similar in the two groups (25.8% with nirmatrelvir–ritonavir and 24.1% with placebo). In the nirmatrelvir–ritonavir group, the most commonly reported treatment-related adverse events were dysgeusia (in 5.8% of the participants) and diarrhea (in 2.1%). CONCLUSIONS The time to sustained alleviation of all signs and symptoms of Covid-19 did not differ significantly between participants who received nirmatrelvir–ritonavir and those who received placebo. (Supported by Pfizer; EPIC-SR ClinicalTrials.gov number, NCT05011513.) Published in NEJM (April 3, 2024):
|



 Your new post is loading...
Your new post is loading...


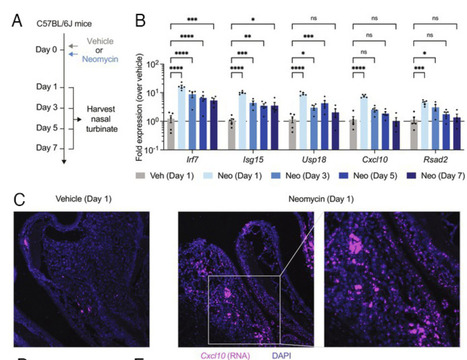

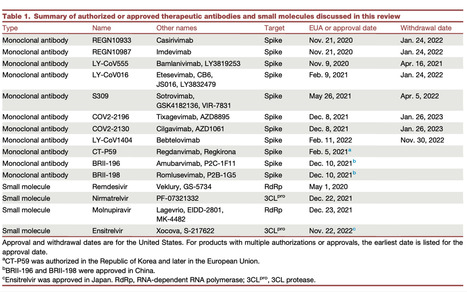


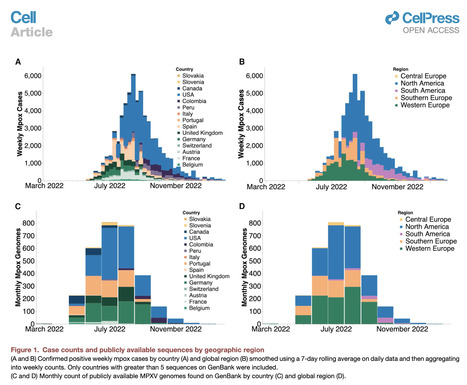



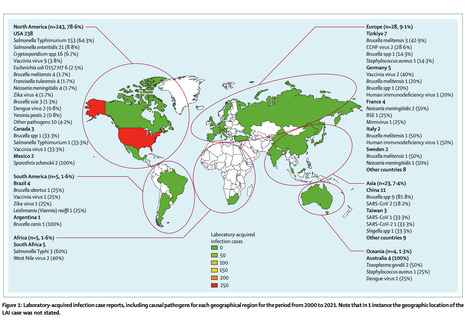
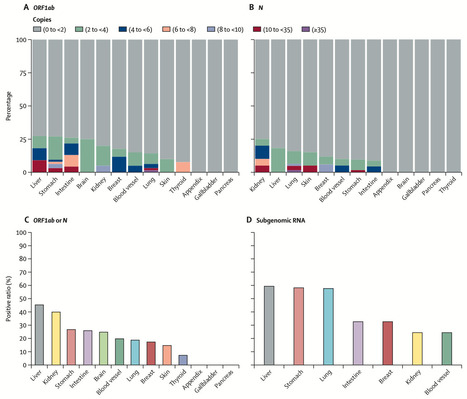


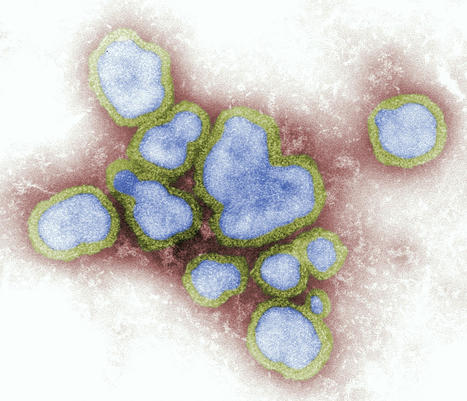


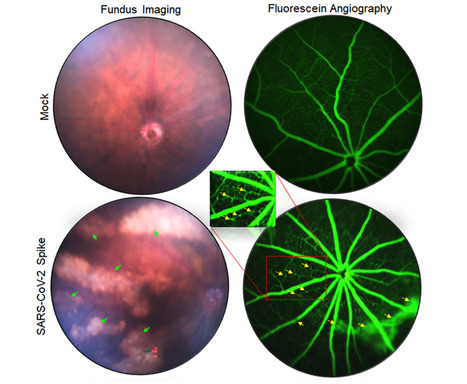
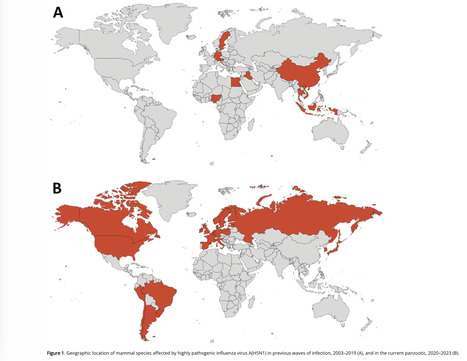







Buy Klonopin without prescription
buy liquid morphine online
Buy Morphine online uk
Buy Norco Online Without Prescription
Buy Oramorph without prescription
Buy oxycontin online uk
Buy Quaaludes Online
buy sildenafil 50 mg tablet
Buy yellow Xanax online
Dilaudid for pain
Order Acxion phentermine 30mg
Order codeine without prescription