Wake Forest Institute for Regenerative Medicine (WFIRM) scientists working on CRISPR/Cas9-mediated gene editing technology have developed a method to increase efficiency of editing while minimizing DNA deletion sizes, a key step toward developing gene editing therapies to treat genetic diseases.
Research and publish the best content.
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
|
|



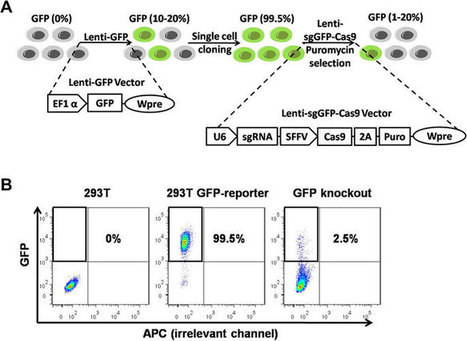






Although CRISPR/Cas9 mainly generates short insertions or deletions at the target site, it can also make large deletions of DNA around the specific target site. These large deletions pose safety concerns and can reduce the efficiency of functional editing. The WFIRM team is looking at ways to reduce the risk of this happening. The research described in their recent paper, published recently in Nucleic Acids Research, aimed to combat the generation of unpredictable long DNA deletions on target and find a way to prevent this, a key step towards the development of gene editing therapies to treat genetic diseases. The team evaluated a variety of human cells and genes of interest and found that fusing DNA polymerase I or the Klenow fragment to the Cas9 enzyme minimised large, unanticipated deletions of genomic DNA without sacrificing the efficiency of genome editing. On the contrary, it even increased editing efficiency in primary human cells.